
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,359,684 – 11,359,924 |
| Length | 240 |
| Max. P | 0.999790 |

| Location | 11,359,684 – 11,359,804 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -43.20 |
| Consensus MFE | -38.94 |
| Energy contribution | -39.90 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11359684 120 - 27905053 GGAACCGCAGACAACAGUGGUUCUGCCCUUCGAGGGCGGCUUGCAGGUGUACGCUGCCACCCAGUGGAUGGACCUAACCCAGGACACCAUCGCCAAUGUGCUGAACCUCAAGUCCAACGA ((((((((........))))))))((((.....))))((((((.((((((((((((.....)))).(((((.(((.....)))...)))))......))))...))))))))))...... ( -44.50) >DroSec_CAF1 107232 120 - 1 GGAACCGCAGACAACAGUGGUUCUGCCCUUCGAGGGCGGCUUGCAGGUGUACGCUGCCACCCAGUGGAUGGACCUAACCCAGGACACCAUCGCCAAUGUGCUGAACCUCAAGUCCAACGA ((((((((........))))))))((((.....))))((((((.((((((((((((.....)))).(((((.(((.....)))...)))))......))))...))))))))))...... ( -44.50) >DroSim_CAF1 98014 120 - 1 GGAACCGCAGACAACAGUGGUUCUGCCUUUCGAGGGCGGCUUGCAGGUGUACACUGCCACCCAGUGGAUGGACCUAACCCAGGACACCAUCGCCAAUGUGCUGAACCUCAAGUCCAACGA ((((((((........))))))))((((.....))))((((((.((((...(((((.....)))))(((((.(((.....)))...))))).............))))))))))...... ( -42.10) >DroEre_CAF1 94662 120 - 1 GGAACCGCAGACGACAGUGGUUCUGCCCUUCGAGGGCGGCCUGCAGGUCUACUCGGCCACUCAGUGGAUGGACCUCACCCAGGACACCAUCGCGAAUGUGCUGAACCUCAAGUCCAACGA ((((((((........))))))))((((.....))))(((.((.((((....(((((((.((.(((.((((.(((.....)))...))))))))).)).))))))))))).)))...... ( -42.80) >DroYak_CAF1 97337 120 - 1 GGAACCGCAGACAACCGUGGUUGUGCCCUUCGAGGGCGGCCUGCAGGUGUACUCGGCCACCCAGUGGAUGGACCUAACCCAGGACACCAUCGCCAAUGUGCUGAACCUCAAGUCCAACGA (((...(((((((((....)))))((((.....))))...))))((((((((..((((((...)))(((((.(((.....)))...))))))))...))))...))))....)))..... ( -42.10) >consensus GGAACCGCAGACAACAGUGGUUCUGCCCUUCGAGGGCGGCUUGCAGGUGUACGCUGCCACCCAGUGGAUGGACCUAACCCAGGACACCAUCGCCAAUGUGCUGAACCUCAAGUCCAACGA ((((((((........))))))))((((.....))))((((((.(((((((((..(((((...)))(((((.(((.....)))...)))))))...)))))...))))))))))...... (-38.94 = -39.90 + 0.96)

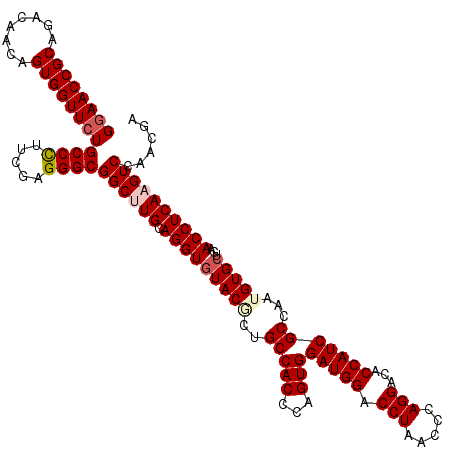
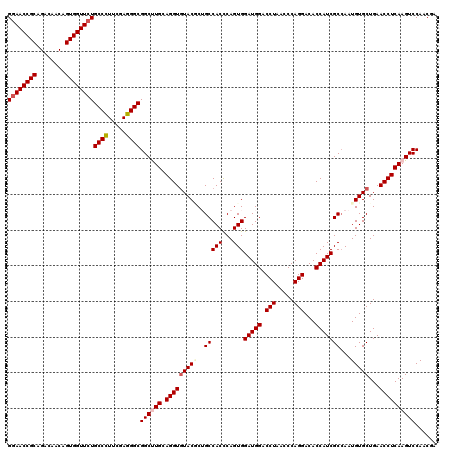
| Location | 11,359,724 – 11,359,844 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -46.04 |
| Consensus MFE | -38.52 |
| Energy contribution | -38.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11359724 120 - 27905053 CCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAUGGAACCGCAGACAACAGUGGUUCUGCCCUUCGAGGGCGGCUUGCAGGUGUACGCUGCCACCCAGUGGAUGGACCUAACCC ..(((((((((((........)))))).(((((.......((((((((........)))))))).......(..((((((.(((....))).))))))..).))))).))).))...... ( -45.70) >DroSec_CAF1 107272 120 - 1 CCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAUGGAACCGCAGACAACAGUGGUUCUGCCCUUCGAGGGCGGCUUGCAGGUGUACGCUGCCACCCAGUGGAUGGACCUAACCC ..(((((((((((........)))))).(((((.......((((((((........)))))))).......(..((((((.(((....))).))))))..).))))).))).))...... ( -45.70) >DroSim_CAF1 98054 120 - 1 CCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAUGGAACCGCAGACAACAGUGGUUCUGCCUUUCGAGGGCGGCUUGCAGGUGUACACUGCCACCCAGUGGAUGGACCUAACCC ..(((((((((((........)))))).(((((.......((((((((........))))))))((((.....))))((...((((.......))))...))))))).))).))...... ( -43.90) >DroEre_CAF1 94702 120 - 1 CCGGUCAGCUGGAUAUGGGACUGCAGUACCACUACUACAUGGAACCGCAGACGACAGUGGUUCUGCCCUUCGAGGGCGGCCUGCAGGUCUACUCGGCCACUCAGUGGAUGGACCUCACCC ((((....))))....(((..(((((..((..........((((((((........))))))))((((.....)))))).)))))((((((.((.((......)).))))))))...))) ( -46.50) >DroYak_CAF1 97377 120 - 1 CCGGUCAGCUGGAUAUGGGUCUGCAGUACCACUACUACAUGGAACCGCAGACAACCGUGGUUGUGCCCUUCGAGGGCGGCCUGCAGGUGUACUCGGCCACCCAGUGGAUGGACCUAACCC ..((((.(((((....((((((((.((.(((........))).)).))))))..))((((((((((((.....)))))(((....))).....)))))))))))).....))))...... ( -48.40) >consensus CCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAUGGAACCGCAGACAACAGUGGUUCUGCCCUUCGAGGGCGGCUUGCAGGUGUACGCUGCCACCCAGUGGAUGGACCUAACCC ..(((((((((((........)))))).(((((.......((((((((........))))))))((((.....))))((...(((((....).))))...))))))).))).))...... (-38.52 = -38.72 + 0.20)



| Location | 11,359,764 – 11,359,884 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -52.58 |
| Consensus MFE | -46.46 |
| Energy contribution | -47.18 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11359764 120 + 27905053 GCCGCCCUCGAAGGGCAGAACCACUGUUGUCUGCGGUUCCAUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCAAAGGGUUCCUCCAGCUGGAGGACAUC .((((((.....))))((.(((((((....(((((......)))))))))))).)))).((((....(((((((....((((((((((........)))))))))))))))))))))... ( -53.30) >DroSec_CAF1 107312 120 + 1 GCCGCCCUCGAAGGGCAGAACCACUGUUGUCUGCGGUUCCAUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCGAAGGGUUCCUCUAGCUGGAGGACCUC .((((((.....))))((.(((((((....(((((......)))))))))))).)))).((((....(((((((....((((((((((........)))))))))))))))))))))... ( -50.70) >DroSim_CAF1 98094 120 + 1 GCCGCCCUCGAAAGGCAGAACCACUGUUGUCUGCGGUUCCAUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCGAAAGGUUCCUCUAGCUGGAGGACCUC ...((((((....(((...(((((((....(((((......))))))))))))((((((........))))))..))).)))).))..........((((.(((((....))))))))). ( -43.90) >DroEre_CAF1 94742 120 + 1 GCCGCCCUCGAAGGGCAGAACCACUGUCGUCUGCGGUUCCAUGUAGUAGUGGUACUGCAGUCCCAUAUCCAGCUGACCGGAGGAGCUCACAUCGAAGGGUUCCUCCAGCUGGAGGACAUC ...((((.....))))........(((((.(((((((.((((......)))).))))))).).....(((((((....((((((((((........))))))))))))))))).)))).. ( -56.30) >DroYak_CAF1 97417 120 + 1 GCCGCCCUCGAAGGGCACAACCACGGUUGUCUGCGGUUCCAUGUAGUAGUGGUACUGCAGACCCAUAUCCAGCUGACCGGAGGAGCUCACGUCGAAGGGUUCCUCCAGCUGGAGCACAUC ((.((((.....))))........((..(((((((((.((((......)))).)))))))))))...(((((((....((((((((((........)))))))))))))))))))..... ( -58.70) >consensus GCCGCCCUCGAAGGGCAGAACCACUGUUGUCUGCGGUUCCAUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCGAAGGGUUCCUCCAGCUGGAGGACAUC ...((((.....))))((.(((((((....(((((......)))))))))))).))...((((....(((((((....((((((((((........)))))))))))))))))))))... (-46.46 = -47.18 + 0.72)

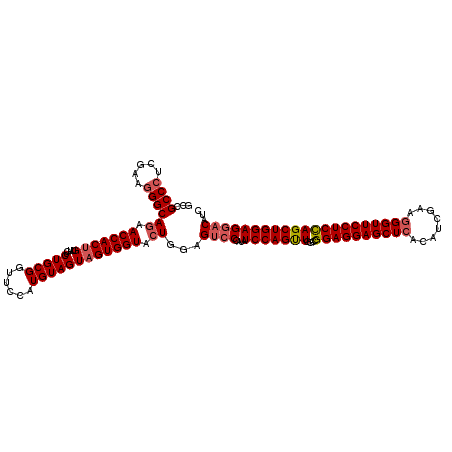
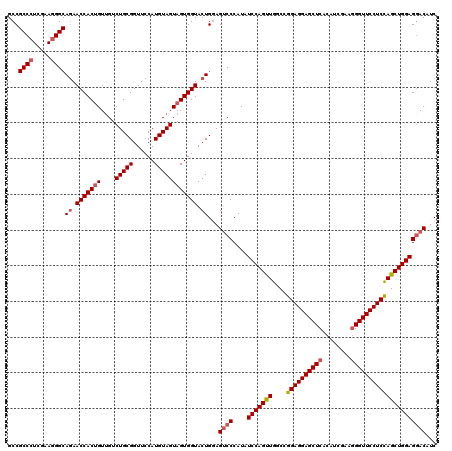
| Location | 11,359,804 – 11,359,924 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -49.18 |
| Consensus MFE | -42.32 |
| Energy contribution | -42.60 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11359804 120 + 27905053 AUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCAAAGGGUUCCUCCAGCUGGAGGACAUCGAUGGUGGACUUGAUGGGCUGCUCCAACCGCUUGUUAUUG .......((((((..(((((((((((.(((((((....((((((((((........)))))))))))))))))(..(((.....)))..)...)))))..))))))))))))........ ( -51.60) >DroSec_CAF1 107352 120 + 1 AUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCGAAGGGUUCCUCUAGCUGGAGGACCUCGAUGGUGGACUUGAUGGGCUGCUCCAACCGCUUGUUAUUG .......((((((..(((((((((((.(((..(..((..(((((((((........)))))))))..))..)...(((.....))))))....)))))..))))))))))))........ ( -49.80) >DroSim_CAF1 98134 120 + 1 AUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCGAAAGGUUCCUCUAGCUGGAGGACCUCGAUGGUGGACUUGAUGGGCUGCUCCAACCGCUUGUUAUUG .......((((((..(((((((((((....((((.((((((((.......(((....)))((((((....))))))))))..)))).))))..)))))..))))))))))))........ ( -47.60) >DroEre_CAF1 94782 120 + 1 AUGUAGUAGUGGUACUGCAGUCCCAUAUCCAGCUGACCGGAGGAGCUCACAUCGAAGGGUUCCUCCAGCUGGAGGACAUCGAUGGAGGACUUGACGGGUUGCUGGAACCGCUUGUUGUUU .......((((((.((((((.(((...(((((((....((((((((((........)))))))))))))))))(..(.((....)))..).....))))))).)).))))))........ ( -47.40) >DroYak_CAF1 97457 120 + 1 AUGUAGUAGUGGUACUGCAGACCCAUAUCCAGCUGACCGGAGGAGCUCACGUCGAAGGGUUCCUCCAGCUGGAGCACAUCGAUGGUGGACUUGAUGGCUUGCUCCAACCGCUUGCUGUUG ...((((((((((...((((.......(((((((....((((((((((........)))))))))))))))))((.((((((.(.....)))))))))))))....))))).)))))... ( -49.50) >consensus AUGUAGUAGUGGUACUGGAGUCCCAUAUCCAGUUGGCCGGAGGAGCUCACAUCGAAGGGUUCCUCCAGCUGGAGGACAUCGAUGGUGGACUUGAUGGGCUGCUCCAACCGCUUGUUAUUG .......((((((..(((((((((...(((((((....((((((((((........))))))))))))))))).....((((.(.....))))).)))..))))))))))))........ (-42.32 = -42.60 + 0.28)

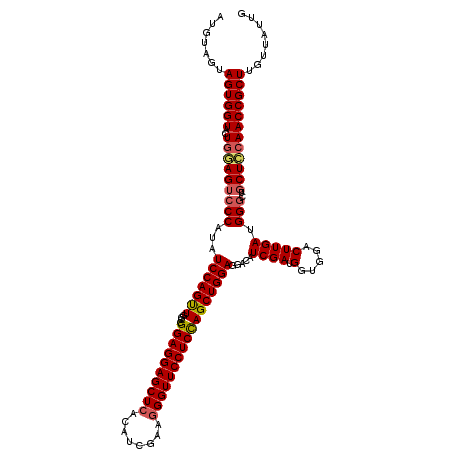

| Location | 11,359,804 – 11,359,924 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -41.54 |
| Consensus MFE | -34.58 |
| Energy contribution | -35.62 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11359804 120 - 27905053 CAAUAACAAGCGGUUGGAGCAGCCCAUCAAGUCCACCAUCGAUGUCCUCCAGCUGGAGGAACCCUUUGAUGUGAGCUCCUCCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAU ...........((((((((.(((((((((((((.......))).((((((....)))))).....)))))).).))).)))))))).((((((........))))))............. ( -42.20) >DroSec_CAF1 107352 120 - 1 CAAUAACAAGCGGUUGGAGCAGCCCAUCAAGUCCACCAUCGAGGUCCUCCAGCUAGAGGAACCCUUCGAUGUGAGCUCCUCCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAU ........((.((((((((...(((((..((..(((.((((((((((((......)))))...))))))))))..))..(((((....))))).))))).)))))..))).))....... ( -42.60) >DroSim_CAF1 98134 120 - 1 CAAUAACAAGCGGUUGGAGCAGCCCAUCAAGUCCACCAUCGAGGUCCUCCAGCUAGAGGAACCUUUCGAUGUGAGCUCCUCCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAU ........((.((((((((...(((((..((..(((.((((((((((((......)))).)))..))))))))..))..(((((....))))).))))).)))))..))).))....... ( -42.00) >DroEre_CAF1 94782 120 - 1 AAACAACAAGCGGUUCCAGCAACCCGUCAAGUCCUCCAUCGAUGUCCUCCAGCUGGAGGAACCCUUCGAUGUGAGCUCCUCCGGUCAGCUGGAUAUGGGACUGCAGUACCACUACUACAU .........(((((((((............(..(((((((((..((((((....)))))).....)))))).)))..).(((((....)))))..)))))))))((((....)))).... ( -43.50) >DroYak_CAF1 97457 120 - 1 CAACAGCAAGCGGUUGGAGCAAGCCAUCAAGUCCACCAUCGAUGUGCUCCAGCUGGAGGAACCCUUCGACGUGAGCUCCUCCGGUCAGCUGGAUAUGGGUCUGCAGUACCACUACUACAU .....((...(((((((((((...((((............)))))))))))))))..(((.(((...(((....).)).(((((....)))))...))))))))((((....)))).... ( -37.40) >consensus CAAUAACAAGCGGUUGGAGCAGCCCAUCAAGUCCACCAUCGAUGUCCUCCAGCUGGAGGAACCCUUCGAUGUGAGCUCCUCCGGCCAACUGGAUAUGGGACUCCAGUACCACUACUACAU ........((.((((((((...(((((..((..(((.(((((..((((((....)))))).....))))))))..))..(((((....))))).))))).)))))..))).))....... (-34.58 = -35.62 + 1.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:22:04 2006