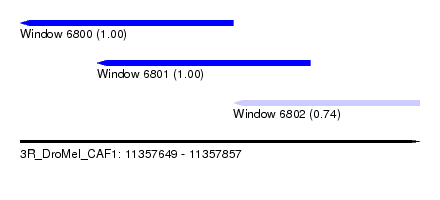
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,357,649 – 11,357,857 |
| Length | 208 |
| Max. P | 0.999985 |

| Location | 11,357,649 – 11,357,760 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.84 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -28.46 |
| Energy contribution | -28.22 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.37 |
| SVM RNA-class probability | 0.999985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11357649 111 - 27905053 U-------UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA--GAGGUCUUCAUUAAUUUUAACAUAAAUUGUUGUAUAGAAAUUUUAUGCCACACGAAGCGUUAAGUGU .-------.(((((((((((...(((((.....((((.......))))...--)))))......................(((.(((((((...))))))).)))..))))))))))).. ( -29.60) >DroSec_CAF1 105197 111 - 1 U-------UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA--GAGGUCUUCAUUAAUUUUAACAUAAAUUGUUGUAUAGAAAUUUUAUGCCACACGGAGCGUUAAGUGU .-------.(((((((((((...(((((.....((((.......))))...--)))))......................(((.(((((((...))))))).)))..))))))))))).. ( -30.00) >DroSim_CAF1 95979 111 - 1 U-------UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA--GAGGUCUUCAUUAAUUUUAACAUAAAUUGUUGUAUAGAAAUUUUAUGCCACACGGAGCGUUAAGUGU .-------.(((((((((((...(((((.....((((.......))))...--)))))......................(((.(((((((...))))))).)))..))))))))))).. ( -30.00) >DroEre_CAF1 92622 111 - 1 C-------CGCUUAAUGCUUAACACCUCAACACGCGGACCCACCCCGCCAA--GAGCUCUUCAUUAAUUUUAACAUAAAUUGUUGUAUAGAAAUUUUAUGCCACGCGGAGCGUUAAGUGU .-------((((((((((((.............((((.......))))...--..((........(((((......)))))((.(((((((...))))))).)))).)))))))))))). ( -27.60) >DroYak_CAF1 95285 120 - 1 UUGCUUAACGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCGAAACGAGCUCUUCAUUAAUUUCCACAUAAAUUGUUGUAUAGAAAUGUUAUGCCACGCGGAGCGUUAAGUGU ..(((((((((((...((.(((((.(((....(((((.......)))))....))).(((.((.((((((......)))))).))...)))..))))).))......))))))))))).. ( -33.20) >consensus U_______UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA__GAGGUCUUCAUUAAUUUUAACAUAAAUUGUUGUAUAGAAAUUUUAUGCCACACGGAGCGUUAAGUGU ........((((((((((((...(((((.....((((.......)))).....)))))......................(((.((((((.....)))))).)))..)))))))))))). (-28.46 = -28.22 + -0.24)

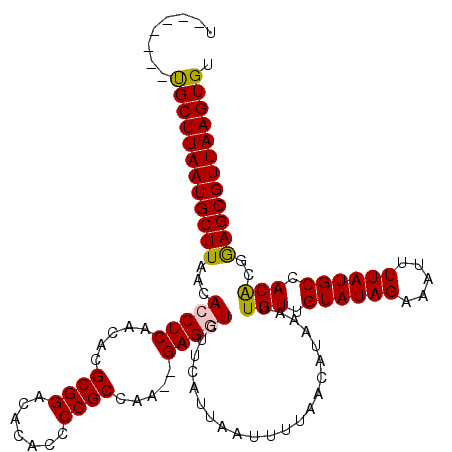
| Location | 11,357,689 – 11,357,800 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -23.76 |
| Energy contribution | -23.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11357689 111 - 27905053 CCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUCU-------UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA--GAGGUCUUCAUUAAUUUUAACAUAAAU ...((((...((((((((((((........)))))))....-------)))))...))))...(((((.....((((.......))))...--)))))...................... ( -26.70) >DroSec_CAF1 105237 111 - 1 CCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUCU-------UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA--GAGGUCUUCAUUAAUUUUAACAUAAAU ...((((...((((((((((((........)))))))....-------)))))...))))...(((((.....((((.......))))...--)))))...................... ( -26.70) >DroSim_CAF1 96019 111 - 1 CCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUCU-------UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA--GAGGUCUUCAUUAAUUUUAACAUAAAU ...((((...((((((((((((........)))))))....-------)))))...))))...(((((.....((((.......))))...--)))))...................... ( -26.70) >DroEre_CAF1 92662 111 - 1 CCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUCC-------CGCUUAAUGCUUAACACCUCAACACGCGGACCCACCCCGCCAA--GAGCUCUUCAUUAAUUUUAACAUAAAU ..(((((((..(((....((((........))))(((...(-------(((......................)))).))).....)))..--))))))).................... ( -30.45) >DroYak_CAF1 95325 120 - 1 CCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUCUUGCUUAACGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCGAAACGAGCUCUUCAUUAAUUUCCACAUAAAU ..(((((((.((((((((((((........)))))(((....)))........)))))))............(((((.......)))))....))))))).................... ( -33.30) >consensus CCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUCU_______UGCUUAAUGCUUAACACCUCAACACGCGGACACACCCCGCCAA__GAGGUCUUCAUUAAUUUUAACAUAAAU ..(((.(((.((((((((((((........)))))(((...........))).))))))).............((((.......)))).....))).))).................... (-23.76 = -23.76 + -0.00)

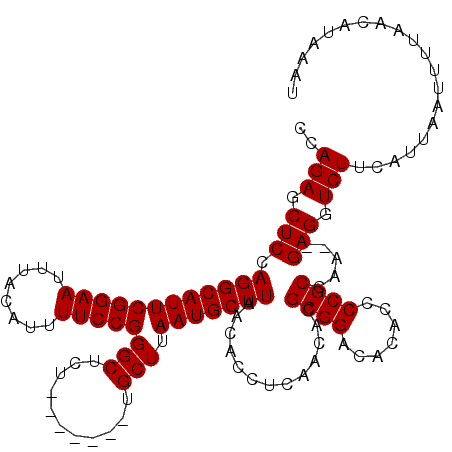
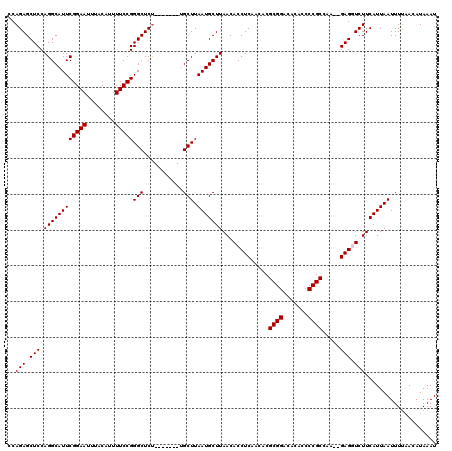
| Location | 11,357,760 – 11,357,857 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.07 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11357760 97 - 27905053 AUCCAAAUGACAUUCAUUGCCAGCCAGCCGUCGGCCUGGGUCAACAGAAGCCC-------CUGUCCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUC .....((((.....))))...((((.(((...(((((((....((((......-------)))))))).)))...)))...(((((........))))))))). ( -30.70) >DroSec_CAF1 105308 97 - 1 AUCCAAAUGACAUUCAUUGCCAGCCAGCCGUCGGCCUGGGUCAACAGAAGCCC-------CUGUCCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUC .....((((.....))))...((((.(((...(((((((....((((......-------)))))))).)))...)))...(((((........))))))))). ( -30.70) >DroSim_CAF1 96090 97 - 1 AUCCAAAUGACAUUCAUUGCCAGCCAGUCGUCGGCCUGGGUCAACAGAAGCCC-------CUGUCCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUC .....((((.....))))...((((........((((((((..((((......-------)))).....)).))))))...(((((........))))))))). ( -28.10) >DroEre_CAF1 92733 104 - 1 AUCCGAAUGACAUUCAUUGCCAGCCAGCCGUCGGCCUGGGUCAACAGAAGCCCCUGGCCCCUGUCCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUC .....((((.....))))...((((.(((...(((((((....((((..(((...)))..)))))))).)))...)))...(((((........))))))))). ( -34.70) >consensus AUCCAAAUGACAUUCAUUGCCAGCCAGCCGUCGGCCUGGGUCAACAGAAGCCC_______CUGUCCAGAGCUCCAGGCAUUCGGAAUUUACAUUUUCCGGGCUC .....((((.....))))...((((.(((...(((((((....((((.............)))))))).)))...)))...(((((........))))))))). (-28.26 = -28.07 + -0.19)

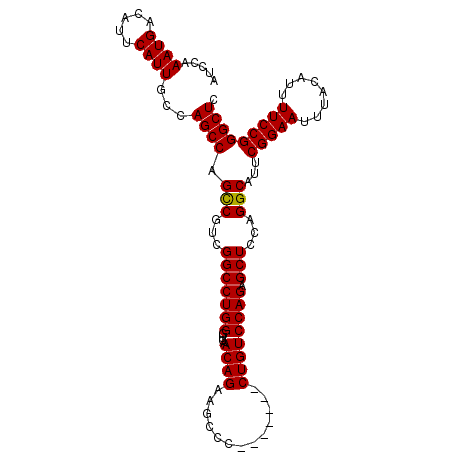
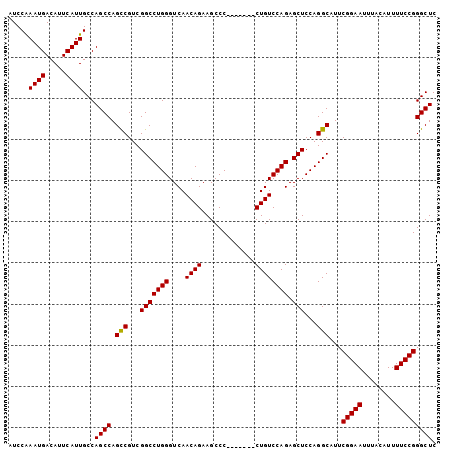
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:54 2006