
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,354,677 – 11,354,913 |
| Length | 236 |
| Max. P | 0.889533 |

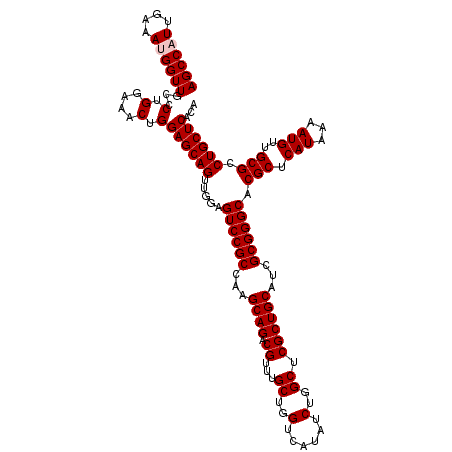
| Location | 11,354,677 – 11,354,797 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -43.94 |
| Consensus MFE | -41.34 |
| Energy contribution | -41.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.889533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11354677 120 + 27905053 AGCCAUUGAAAUGGUUGCCCGGGAAACUGGAGCAGUUGGAGUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACA ((((((....))))))...(((....)))((((((.....((((((...((((.((...((..(......)..)).))))))...)))))).(((.(((...)))..))).))))))... ( -47.00) >DroSec_CAF1 102245 120 + 1 AGCCAUUGAAAUGGUUGCCCUGGAAACUGGAGCAGUUGGAGUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACA ((((((....)))))).....(....)..((((((.....((((((...((((.((...((..(......)..)).))))))...)))))).(((.(((...)))..))).))))))... ( -43.10) >DroSim_CAF1 93020 120 + 1 AGCCAUUGAAAUGGUUGCCCUGGAAACUGGAGCAGUUGGAGUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACA ((((((....)))))).....(....)..((((((.....((((((...((((.((...((..(......)..)).))))))...)))))).(((.(((...)))..))).))))))... ( -43.10) >DroEre_CAF1 89782 120 + 1 AGCCAUUGAAAAGGUUUCCCUGGAAACUGGAGCAGUUGGAGUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACA ............((((((....)))))).((((((.....((((((...((((.((...((..(......)..)).))))))...)))))).(((.(((...)))..))).))))))... ( -43.90) >DroYak_CAF1 92279 120 + 1 AGCCAUUGAAAAGGUUGCACUGGAAACUGGAGCAGUUGGAGUCCGCCGAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACA ((((........))))...(.(....).)((((((.....((((((.((((((.((...((..(......)..)).)))))).)))))))).(((.(((...)))..))).))))))... ( -42.60) >consensus AGCCAUUGAAAUGGUUGCCCUGGAAACUGGAGCAGUUGGAGUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACA ((((((....))))))...(.(....).)((((((.....((((((...((((.((...((..(......)..)).))))))...)))))).(((.(((...)))..))).))))))... (-41.34 = -41.74 + 0.40)


| Location | 11,354,717 – 11,354,837 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.49 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -33.44 |
| Energy contribution | -33.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11354717 120 + 27905053 GUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAACUCCACUCCACUAUCCCUC ((((((...((((.((...((..(......)..)).))))))...)))))).............((((((((((........))))..........)))))).................. ( -33.20) >DroSec_CAF1 102285 120 + 1 GUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAACUCCACUCCAACACCCAUC ((((((...((((.((...((..(......)..)).))))))...))))))............(((((.(..((........(((((......)))))......))..))))))...... ( -33.34) >DroSim_CAF1 93060 120 + 1 GUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCGGAUCCUGCAACUCCACUCCACCACCCAUC ((((((...((((.((...((..(......)..)).))))))...)))))).............(((((.((((........))))....(....).))))).................. ( -34.40) >DroEre_CAF1 89822 107 + 1 GUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAGCUCCUC------------- ((((((...((((.((...((..(......)..)).))))))...))))))..(((.......(((.((....))..)))..(((((......))))).))).....------------- ( -33.60) >DroYak_CAF1 92319 118 + 1 GUCCGCCGAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAGCUCCACUCCU--UCCCAUC ((((((.((((((.((...((..(......)..)).)))))).))))))))..(((.......(((.((....))..)))..(((((......))))).))).........--....... ( -34.90) >consensus GUCCGCCAAGCAGACGUUUGCUGGUCAUAUCUGGCUCGCUGCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAACUCCACUCCA__ACCCAUC ((((((...((((.((...((..(......)..)).))))))...)))))).............((((((((((........))))..........)))))).................. (-33.44 = -33.20 + -0.24)



| Location | 11,354,757 – 11,354,875 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -22.32 |
| Energy contribution | -23.56 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11354757 118 + 27905053 GCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAACUCCACUCCACUAUCCCUC-CCCAUCUCCAUCCCCU-CUCUUGGGUCCUGGGUCGACAA ...(((.(((((.((.((........)).)).)))))..((((((((..(((...)))......................-............(((.-.....))))))))))))))... ( -29.60) >DroSec_CAF1 102325 118 + 1 GCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAACUCCACUCCAACACCCAUC-CCCAUCUCCAUCCCCU-CUCUUGGGUCCUGGGUCGACAA ...(((.(((((.((.((........)).)).)))))..((((((((..(((...)))......................-............(((.-.....))))))))))))))... ( -29.60) >DroSim_CAF1 93100 118 + 1 GCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCGGAUCCUGCAACUCCACUCCACCACCCAUC-CCCAUCUCCAUCCCCU-CUCUUGGGUCCUGGGUCGACAA ...(((.(((((.((.((........)).)).)))))..((((((((...(((.........)))...............-............(((.-.....))))))))))))))... ( -32.20) >DroEre_CAF1 89862 91 + 1 GCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAGCUCCUC-------------------------CCCACCCCUC----UCCUGGGUCGACAA ...(((.(((((.((.((........)).)).)))))..((((((((....................-------------------------..........----)))))))))))... ( -23.41) >DroYak_CAF1 92359 113 + 1 GCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAGCUCCACUCCU--UCCCAUCUCUCAUAUCCACCCCAC-CCCC----UCCUGAGUCGACAA ((...((((((.((..(((...))).)).)))))).......(((((......)))))..))((.((((..--........................-....----....)))).))... ( -20.28) >consensus GCAUCGCGGGCACGCUCAUAAAAUGUUGCGCCUGCUCACACCCAGGAAUCAGAUCCUGCAACUCCACUCCA__ACCCAUC_CCCAUCUCCAUCCCCU_CUCUUGGGUCCUGGGUCGACAA ...(((.(((((.((.((........)).)).)))))..((((((((..(((...)))...................................(((.......))))))))))))))... (-22.32 = -23.56 + 1.24)



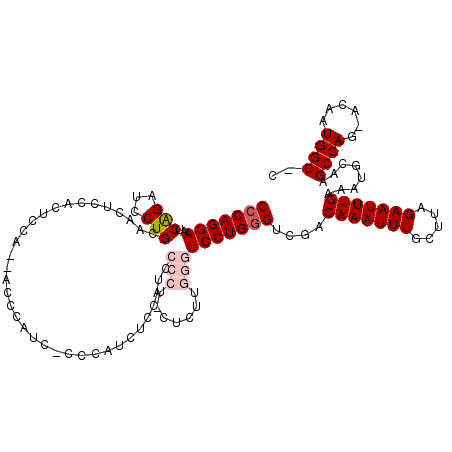
| Location | 11,354,797 – 11,354,913 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.92 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11354797 116 + 27905053 CCCAGGAAUCAGAUCCUGCAACUCCACUCCACUAUCCCUC-CCCAUCUCCAUCCCCU-CUCUUGGGUCCUGGGUCGACAAAUUUGCUUAGAAUUUGAAAUGCAAGCCAG-ACAAUGGC-C (((((((..(((...)))......................-............(((.-.....))))))))))....(((((((.....)))))))........((((.-....))))-. ( -25.40) >DroSec_CAF1 102365 118 + 1 CCCAGGAAUCAGAUCCUGCAACUCCACUCCAACACCCAUC-CCCAUCUCCAUCCCCU-CUCUUGGGUCCUGGGUCGACAAAUUUGCUUAGAAUUUGAAAUGCAAGCCAGCACAAUGGCCC ...((((......))))(((.............(((((..-((((............-....))))...)))))...(((((((.....)))))))...)))..((((......)))).. ( -26.49) >DroSim_CAF1 93140 116 + 1 CCCAGGAAUCGGAUCCUGCAACUCCACUCCACCACCCAUC-CCCAUCUCCAUCCCCU-CUCUUGGGUCCUGGGUCGACAAAUUUGCUUAGAAUUUGAAAUGCAAGCCAG-ACAAUGGC-C (((((((...(((.........)))...............-............(((.-.....))))))))))....(((((((.....)))))))........((((.-....))))-. ( -28.00) >DroEre_CAF1 89902 89 + 1 CCCAGGAAUCAGAUCCUGCAGCUCCUC-------------------------CCCACCCCUC----UCCUGGGUCGACAAAUUUGCUUAGAAUUUGAAAUGCAAGCCAG-ACAAUGGC-C ..(((((......)))))..((.....-------------------------.....(((..----....)))....(((((((.....)))))))....))..((((.-....))))-. ( -20.80) >DroYak_CAF1 92399 111 + 1 CCCAGGAAUCAGAUCCUGCAGCUCCACUCCU--UCCCAUCUCUCAUAUCCACCCCAC-CCCC----UCCUGAGUCGACAAAUUUGCUUAGAAUUUGAAAUGCAAGCCAG-ACAAUGGC-C ..(((((......)))))..((((.((((..--........................-....----....)))).))(((((((.....)))))))....))..((((.-....))))-. ( -17.78) >consensus CCCAGGAAUCAGAUCCUGCAACUCCACUCCA__ACCCAUC_CCCAUCUCCAUCCCCU_CUCUUGGGUCCUGGGUCGACAAAUUUGCUUAGAAUUUGAAAUGCAAGCCAG_ACAAUGGC_C (((((((..(((...)))...................................(((.......))))))))))....(((((((.....)))))))........((((......)))).. (-17.68 = -18.92 + 1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:49 2006