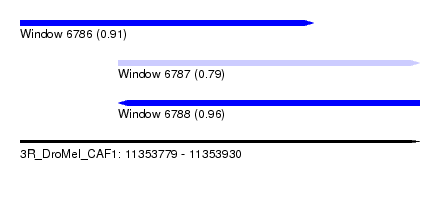
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,353,779 – 11,353,930 |
| Length | 151 |
| Max. P | 0.955069 |

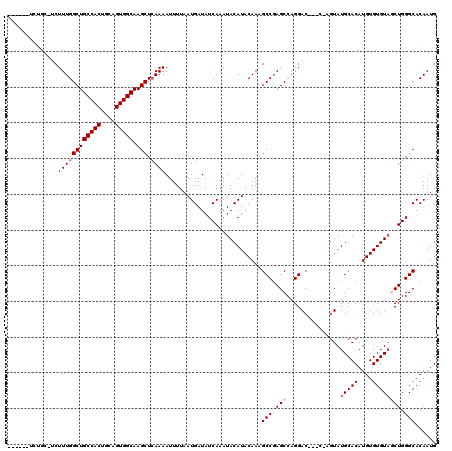
| Location | 11,353,779 – 11,353,890 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.60 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
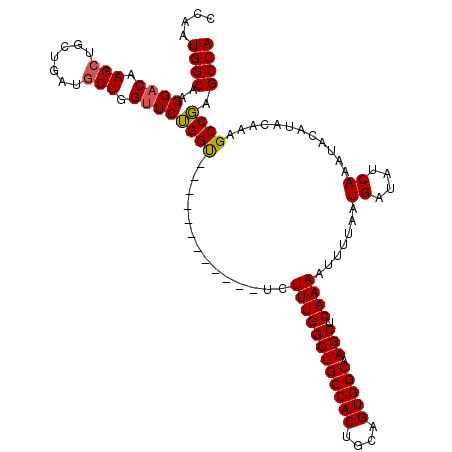
>3R_DroMel_CAF1 11353779 111 + 27905053 CCAAUGGCAAGGACAAGCUGCUGAUGCUGGUUCGGGC------UCUGCAUCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCCAGCCA ....((((..((((.(((.......))).))))((((------(.((....((((((((((((....))))).))).)))).......((....))........)).))))).)))) ( -38.00) >DroSec_CAF1 101342 105 + 1 CCAAUGGCAAGGACAAGCUGCUGAUGCUGGGUCUGGU------------UCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCGAGCCA ....((((..((((.(((.......)))..))))(((------------(.((((((((((((....))))).))).)))).......((....))...........))))..)))) ( -30.80) >DroSim_CAF1 92118 105 + 1 CCAAUGGCAAGGACAAGCUGCUGGUGCUGGUUCUGGU------------UCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCGAGCCA ....((((.(((((.(((.......))).)))))(((------------(.((((((((((((....))))).))).)))).......((....))...........))))..)))) ( -31.30) >DroYak_CAF1 91297 117 + 1 CCAAUGGCAAGGACAAGCUGCUGGUUCUGGUUCUGGUUCUGCUUCUGCUGCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAUGCCGAGCCA (((..(((........)))..)))....(((((.(((...(((((..((((..(((...)))..))))..).))))............((....))............)))))))). ( -32.60) >consensus CCAAUGGCAAGGACAAGCUGCUGAUGCUGGUUCUGGU____________UCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCGAGCCA ....((((..((((.(((.......))).))))((((..............((((((((((((....))))).))).)))).......((....))............)))).)))) (-28.66 = -28.60 + -0.06)

| Location | 11,353,816 – 11,353,930 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -23.82 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11353816 114 + 27905053 ------UCUGCAUCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCCAGCCAGGACGUACCAAUAUGCACAUGUGUGUUGCUGGGCACAAUG ------........((((((((((((....))))).))).))))...........................(((((((..((.....))(((((((....))))))))))))))...... ( -34.70) >DroSec_CAF1 101379 108 + 1 ------------UCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCGAGCCAGGACACACGAGUAUGCACAUGUGUGUAGCUGGGCACAAUG ------------..((((((((((((....))))).))).))))...........................(((.(((....(((((((.((....)).))))))).))).)))...... ( -34.90) >DroSim_CAF1 92155 108 + 1 ------------UCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCGAGCCAGGACAUGCGAGUAUGCACAUGUGUGUAGCUGGGCACAAUG ------------..((((((((((((....))))).))).))))...........................(((.(((..(.(((((.(......).))))).)...))).)))...... ( -32.10) >DroEre_CAF1 88869 114 + 1 GCUGGUUCUGCUUCUUUGGCUGCCACUGCAGUGGCAAGCUGAAAAUUUUAAUGAUAUCAAAUACAUACAAUGCCGAGCCAGGAC------GUAUGCACAUGUGUGUAUCUGGGCACAAUG .(((((((.((...((..((((((((....))))).)))..))........((....))............)).)))))))..(------(.((((((....)))))).))......... ( -35.60) >DroYak_CAF1 91337 111 + 1 ---GCUUCUGCUGCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAUGCCGAGCCAGGAC------GUAUGCACAUGUGUGUAUCUGGGCACAAUG ---((....))(((((((((((((((....))))).))).))))............(((.((((((((((((((......)).)------)).......))))))))).))))))..... ( -30.71) >consensus ______UCUGC_UCUUUGGCUGCCACUGCAGUGGCAAGCUCAAAAUUUUAAUGAUAUCAAAUACAUACAAAGCCGAGCCAGGAC___C_AGUAUGCACAUGUGUGUAGCUGGGCACAAUG ..............((((((((((((....))))).))).))))...........................(((.(((...............(((((....)))))))).)))...... (-23.82 = -24.62 + 0.80)

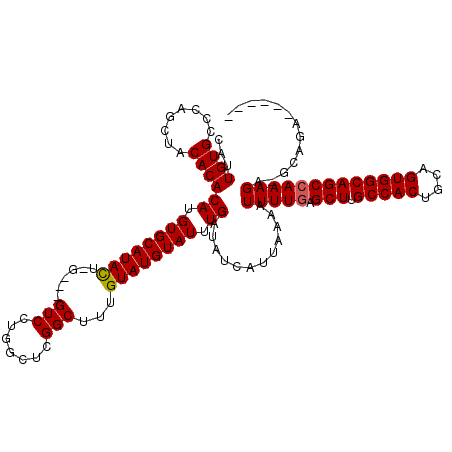
| Location | 11,353,816 – 11,353,930 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.51 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
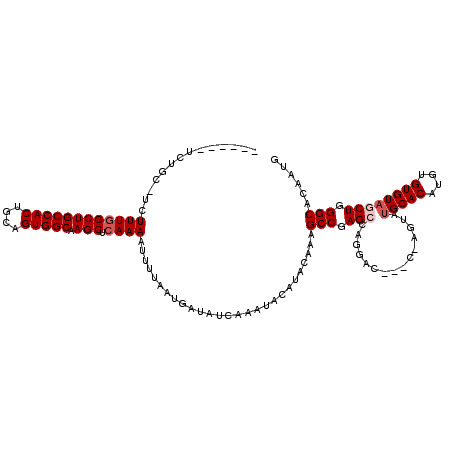
>3R_DroMel_CAF1 11353816 114 - 27905053 CAUUGUGCCCAGCAACACACAUGUGCAUAUUGGUACGUCCUGGCUGGGCUUUGUAUGUAUUUGAUAUCAUUAAAAUUUUGAGCUUGCCACUGCAGUGGCAGCCAAAGAUGCAGA------ ......(((((((.......((((((......))))))....)))))))(((((((...((((((...)))))).(((((.(((.(((((....))))))))))))))))))))------ ( -38.90) >DroSec_CAF1 101379 108 - 1 CAUUGUGCCCAGCUACACACAUGUGCAUACUCGUGUGUCCUGGCUCGGCUUUGUAUGUAUUUGAUAUCAUUAAAAUUUUGAGCUUGCCACUGCAGUGGCAGCCAAAGA------------ ......(((.(((((.(((((((........)))))))..))))).)))..........................(((((.(((.(((((....))))))))))))).------------ ( -34.20) >DroSim_CAF1 92155 108 - 1 CAUUGUGCCCAGCUACACACAUGUGCAUACUCGCAUGUCCUGGCUCGGCUUUGUAUGUAUUUGAUAUCAUUAAAAUUUUGAGCUUGCCACUGCAGUGGCAGCCAAAGA------------ ......(((.(((((...(((((((......)))))))..))))).)))..........................(((((.(((.(((((....))))))))))))).------------ ( -33.70) >DroEre_CAF1 88869 114 - 1 CAUUGUGCCCAGAUACACACAUGUGCAUAC------GUCCUGGCUCGGCAUUGUAUGUAUUUGAUAUCAUUAAAAUUUUCAGCUUGCCACUGCAGUGGCAGCCAAAGAAGCAGAACCAGC ..((.(((.((((((((.(((.((((....------((....))...))))))).))))))))............((((..(((.(((((....))))))))..)))).))).))..... ( -31.70) >DroYak_CAF1 91337 111 - 1 CAUUGUGCCCAGAUACACACAUGUGCAUAC------GUCCUGGCUCGGCAUUGUAUGUAUUUGAUAUCAUUAAAAUUUUGAGCUUGCCACUGCAGUGGCAGCCAAAGCAGCAGAAGC--- ..((.(((.((((((((.(((.((((....------((....))...))))))).))))))))............(((((.(((.(((((....)))))))))))))..))).))..--- ( -34.40) >consensus CAUUGUGCCCAGCUACACACAUGUGCAUACU_G___GUCCUGGCUCGGCUUUGUAUGUAUUUGAUAUCAUUAAAAUUUUGAGCUUGCCACUGCAGUGGCAGCCAAAGA_GCAGA______ ...((((........))))((.((((((((......(((.......)))...)))))))).))............(((((.(((.(((((....)))))))))))))............. (-25.92 = -25.96 + 0.04)

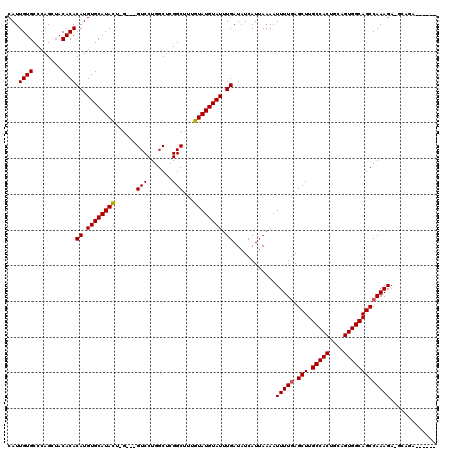
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:41 2006