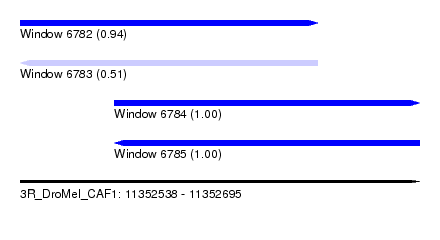
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,352,538 – 11,352,695 |
| Length | 157 |
| Max. P | 0.999851 |

| Location | 11,352,538 – 11,352,655 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -29.12 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11352538 117 + 27905053 -AUAUAUACACAUGCACCCCU-AUCCA-GUCCGAUACUUUCCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAU -...........(((((....-....(-(.((((((..(((...((((((.(((((....)))))...))))))...))).))))))))(((((........)))))...)))))..... ( -30.80) >DroSec_CAF1 100126 117 + 1 -AUAUAUGUACAUGCACCCCU-AUCCA-GUCCGAUACUUUCCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGCGCGAGAAGAUGCGCCCUCUGUGCAGAAAU -...........(((((....-....(-(.((((((..(((...((((((.(((((....)))))...))))))...))).))))))))(((((......))))).....)))))..... ( -27.30) >DroSim_CAF1 90893 117 + 1 -AUAUAUAUAUGUGCACCCCU-AUCCA-GUCCGAUACUUUUCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAU -.........(.(((((....-....(-(.((((((...((((.((((((.(((((....)))))...))))))..)))).))))))))(((((........)))))...))))).)... ( -31.70) >DroEre_CAF1 87660 119 + 1 -AUACAGACAUAUCGACCCCCUAUCCAACCCCGAUAUUUCCCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGCGCGGAAAU -......................(((.....(((((.(((....((((((.(((((....)))))...))))))...))).)))))((.(((((........)))))...))..)))... ( -27.50) >DroYak_CAF1 90066 119 + 1 UAUACAAAUAUAUAGACCCCC-AUCCAAGCCCAAUAUUUUCCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAU ..........((((((.....-.....((((.((((..(((...((((((.(((((....)))))...))))))...))).))))))))(((((........)))))))))))....... ( -28.30) >consensus _AUAUAUAUAUAUGCACCCCU_AUCCA_GUCCGAUACUUUCCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAU ............(((((.............((((((.(((....((((((.(((((....)))))...))))))...))).))))))..(((((........)))))...)))))..... (-23.00 = -23.72 + 0.72)

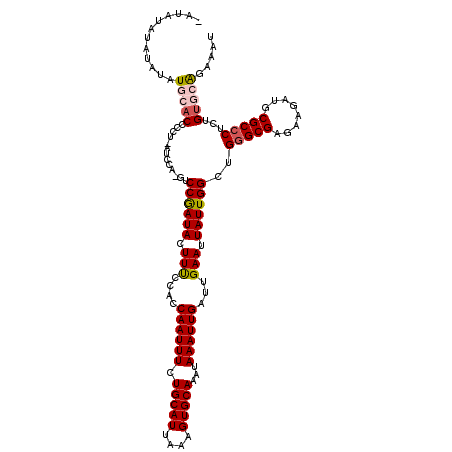

| Location | 11,352,538 – 11,352,655 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.74 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11352538 117 - 27905053 AUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGGAAAGUAUCGGAC-UGGAU-AGGGGUGCAUGUGUAUAUAU- .....((((((....((((..((......((((((.(((.(((.(((((((..((((((......))))))))))))).)))..))).)).)-)))..-))..)))).)))))).....- ( -33.10) >DroSec_CAF1 100126 117 - 1 AUUUCUGCACAGAGGGCGCAUCUUCUCGCGCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGGAAAGUAUCGGAC-UGGAU-AGGGGUGCAUGUACAUAUAU- ((..(((..(((...((((........))))..((.(((.(((.(((((((..((((((......))))))))))))).)))..))).)).)-))..)-))..))..............- ( -29.60) >DroSim_CAF1 90893 117 - 1 AUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGAAAAGUAUCGGAC-UGGAU-AGGGGUGCACAUAUAUAUAU- ((..(((..(((.(((((........)))))..((.(((.((((.((((((..((((((......))))))))))))))))...))).)).)-))..)-))..))..............- ( -31.50) >DroEre_CAF1 87660 119 - 1 AUUUCCGCGCAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGGGAAAUAUCGGGGUUGGAUAGGGGGUCGAUAUGUCUGUAU- ........(((((.(.((.(((((((...((((((.(((.(((.(((((((..((((((......)))))))))))))..))).)))...))))))..)))))))))...).)))))..- ( -36.50) >DroYak_CAF1 90066 119 - 1 AUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGGAAAAUAUUGGGCUUGGAU-GGGGGUCUAUAUAUUUGUAUA ((..(((..(((.(((((........)))))..((((((.(((.(((((((..((((((......))))))))))))).)))..))))))..)))..)-))..))............... ( -30.90) >consensus AUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGGAAAGUAUCGGAC_UGGAU_AGGGGUGCAUAUAUAUAUAU_ ((..((...((..(((((........)))))..((.(((.(((.(((((((..((((((......))))))))))))).)))..))).))...))....))..))............... (-23.88 = -23.72 + -0.16)


| Location | 11,352,575 – 11,352,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -29.18 |
| Energy contribution | -29.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11352575 120 + 27905053 CCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAUUUACUUUUUAAUUUACUCAGAAAACCGAGUAAAAAGGAAA ...(((((((((((((......((.(((((.((....)).))))).)).(((((........)))))...)))))))))))..........(((((((........)))))))..))... ( -32.60) >DroSec_CAF1 100163 120 + 1 CCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGCGCGAGAAGAUGCGCCCUCUGUGCAGAAAUUUACUUUUUAAUUUACUCAGAAAACCGAGUAAAAAGGAAA ...(((((((((((((...((.((.(((((.((....)).))))).)).(((((......))))).))..)))))))))))..........(((((((........)))))))..))... ( -29.30) >DroSim_CAF1 90930 120 + 1 UCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAUUUACUUUUUAAUUUACUCAGAAAACCGAGUAAAAAGGAAA ...(((((((((((((......((.(((((.((....)).))))).)).(((((........)))))...)))))))))))..........(((((((........)))))))..))... ( -32.60) >DroEre_CAF1 87699 120 + 1 CCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGCGCGGAAAUUUACUUUUUAAUUUACUCCGAAAACCGAGUAAAAAGGAAA ...(((((((((((........((.(((((.((....)).))))).)).(((((........))))).....)))))))))..........(((((((........)))))))..))... ( -30.00) >DroYak_CAF1 90105 120 + 1 CCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAUUUACUUUUUAAUUUACUCAGAAAACCGAGCAAAAAGGAAA ...(((((((((((((......((.(((((.((....)).))))).)).(((((........)))))...)))))))))))..........(((.(((........))).)))..))... ( -28.00) >consensus CCACCAAUUUCUGCAUUAAAGUGCAAAUAAAUUGAUUGAAUUAUUGGCUGGGCGAGAAGAUGCGCCCUCUGUGCAGAAAUUUACUUUUUAAUUUACUCAGAAAACCGAGUAAAAAGGAAA ...(((((((((((((......((.(((((.((....)).))))).)).(((((........)))))...)))))))))))..........(((((((........)))))))..))... (-29.18 = -29.62 + 0.44)

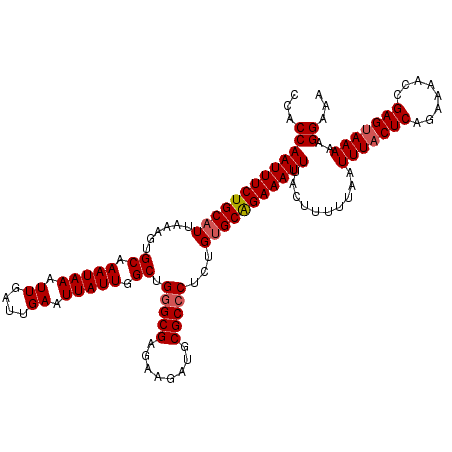
| Location | 11,352,575 – 11,352,695 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -31.12 |
| Energy contribution | -31.40 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11352575 120 - 27905053 UUUCCUUUUUACUCGGUUUUCUGAGUAAAUUAAAAAGUAAAUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGG ...((..(((((((((....)))))))))..........((((((((((....(((((........))))).((.(((((..........))))).)).......))))))))))))... ( -34.40) >DroSec_CAF1 100163 120 - 1 UUUCCUUUUUACUCGGUUUUCUGAGUAAAUUAAAAAGUAAAUUUCUGCACAGAGGGCGCAUCUUCUCGCGCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGG ...((..(((((((((....)))))))))..........((((((((((..((((((((........)))).((.(((((..........))))).)).))))..))))))))))))... ( -35.00) >DroSim_CAF1 90930 120 - 1 UUUCCUUUUUACUCGGUUUUCUGAGUAAAUUAAAAAGUAAAUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGA ...((..(((((((((....)))))))))..........((((((((((....(((((........))))).((.(((((..........))))).)).......))))))))))))... ( -34.40) >DroEre_CAF1 87699 120 - 1 UUUCCUUUUUACUCGGUUUUCGGAGUAAAUUAAAAAGUAAAUUUCCGCGCAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGG .......(((((((.(....).)))))))...............((((.....(((((........)))))...............(((((..((((((......))))))))))))))) ( -27.80) >DroYak_CAF1 90105 120 - 1 UUUCCUUUUUGCUCGGUUUUCUGAGUAAAUUAAAAAGUAAAUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGG ...((..(((((((((....)))))))))..........((((((((((....(((((........))))).((.(((((..........))))).)).......))))))))))))... ( -34.40) >consensus UUUCCUUUUUACUCGGUUUUCUGAGUAAAUUAAAAAGUAAAUUUCUGCACAGAGGGCGCAUCUUCUCGCCCAGCCAAUAAUUCAAUCAAUUUAUUUGCACUUUAAUGCAGAAAUUGGUGG ...((..(((((((((....)))))))))..........((((((((((....(((((........))))).((.(((((..........))))).)).......))))))))))))... (-31.12 = -31.40 + 0.28)

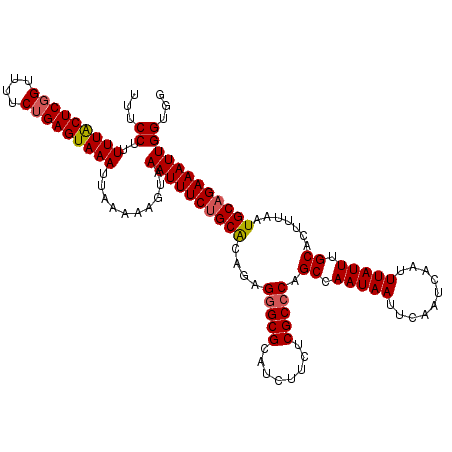
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:38 2006