
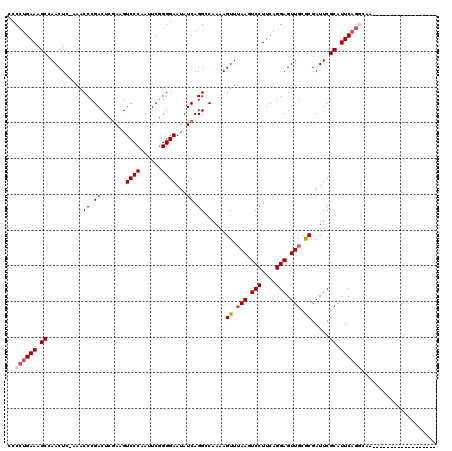
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,350,266 – 11,350,408 |
| Length | 142 |
| Max. P | 0.999908 |

| Location | 11,350,266 – 11,350,386 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
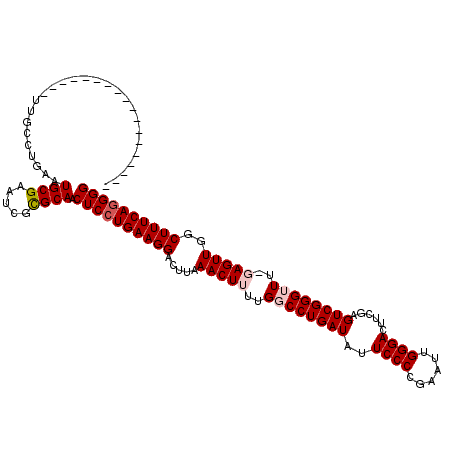
>3R_DroMel_CAF1 11350266 120 + 27905053 GCUUGAUUUAAUUAAUUUCAAUUUUAAAUCAGCUAAAAUCCCCCUGAAAGCCAACUCAAAACCCGACUCGAAGUCCCAAUUCGGGGAAUAUCAGGCCAAAAGUUUAAGUCCUUCAGGAGU (((.(((((((............)))))))))).........((((((.((.((((.....((.((.......((((......))))...)).)).....))))...))..))))))... ( -22.50) >DroSec_CAF1 92502 120 + 1 GCUUGAUUUAAUUAAUUUCAAUUUUAAAUCAGCUAAACUCCCCCUGAAAGCUAACUCCAAACCCGACUCAACGUCCCAAUUCGGGGAAUAUCAGGGCAAAAGUUUAAGUCCUUCAGGAGU (((.(((((((............))))))))))...((((((((((((((....))........(((.....)))....)))))))......(((((..........)))))...))))) ( -27.20) >DroSim_CAF1 87691 120 + 1 GCUUGAUUUAAUUAAUUUCAAUUUUAAAUCAGCUAAAUUCCCCCUGAAAGCCAACUCAAAACCCGACUCGACGUCCCAAUUCGGGGAAUAUCAGGGCAAAAGUUUAAGUCCUUCAGGAGU (((.(((((((............)))))))))).........((((((.....................((..((((......))))...)).((((..........))))))))))... ( -25.40) >DroEre_CAF1 85481 119 + 1 GCGUGAUUUAAUUAAUUUCAAUUUUAAAUCAGCAAAAGUGCCCCUGAAAGCCAACUG-AAACCCGACUUGAAGUCCCAAUUCGGGGAAUAUCAGGCCAAAAGUUUAACUCCUUCAGGAGU ((.((((((((............))))))))))......((.((((((.(((.....-...(((((.(((......))).)))))........)))....((.....))..)))))).)) ( -28.79) >DroYak_CAF1 87780 119 + 1 GCUUGAUUUAAUUAAUUUCAAUUUUAAAUCACCAAAAUUCCCCUUGAAAGCCAACUG-AAACCCGACUUGAAGUCCCAAUUCAGGGAAUAUCAGGCCAAAAGUUGAACUCCUUCAGGAGU (((((((((((............)))))))..(((........))).))))(((((.-......(.(((((..((((......))))...))))))....))))).(((((....))))) ( -26.20) >consensus GCUUGAUUUAAUUAAUUUCAAUUUUAAAUCAGCUAAAUUCCCCCUGAAAGCCAACUC_AAACCCGACUCGAAGUCCCAAUUCGGGGAAUAUCAGGCCAAAAGUUUAAGUCCUUCAGGAGU ((.((((((((............)))))))))).........((((((.(..((((.....((.((.......((((......))))...)).)).....)))).....).))))))... (-20.66 = -20.70 + 0.04)


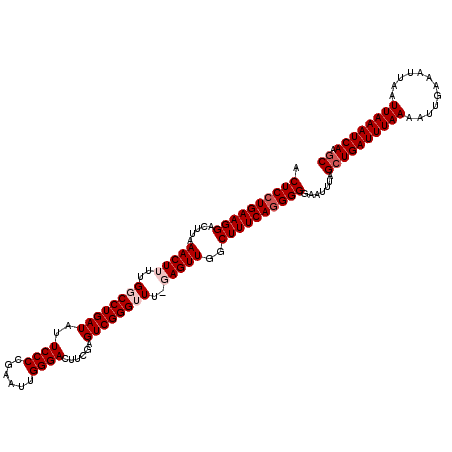
| Location | 11,350,266 – 11,350,386 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.91 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -33.06 |
| Energy contribution | -34.26 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11350266 120 - 27905053 ACUCCUGAAGGACUUAAACUUUUGGCCUGAUAUUCCCCGAAUUGGGACUUCGAGUCGGGUUUUGAGUUGGCUUUCAGGGGGAUUUUAGCUGAUUUAAAAUUGAAAUUAAUUAAAUCAAGC .((((((((((.((..(((((..((((((((..((((......))))......))))))))..))))))))))))))))).......((((((((((............))))))).))) ( -39.60) >DroSec_CAF1 92502 120 - 1 ACUCCUGAAGGACUUAAACUUUUGCCCUGAUAUUCCCCGAAUUGGGACGUUGAGUCGGGUUUGGAGUUAGCUUUCAGGGGGAGUUUAGCUGAUUUAAAAUUGAAAUUAAUUAAAUCAAGC .((((((((((.((..((((((.((((.(((..((((......))))......)))))))..)))))))))))))))))).......((((((((((............))))))).))) ( -36.40) >DroSim_CAF1 87691 120 - 1 ACUCCUGAAGGACUUAAACUUUUGCCCUGAUAUUCCCCGAAUUGGGACGUCGAGUCGGGUUUUGAGUUGGCUUUCAGGGGGAAUUUAGCUGAUUUAAAAUUGAAAUUAAUUAAAUCAAGC .((((((((((.((..(((((..((((((((..((((......)))).))))....))))...))))))))))))))))).......((((((((((............))))))).))) ( -37.00) >DroEre_CAF1 85481 119 - 1 ACUCCUGAAGGAGUUAAACUUUUGGCCUGAUAUUCCCCGAAUUGGGACUUCAAGUCGGGUUU-CAGUUGGCUUUCAGGGGCACUUUUGCUGAUUUAAAAUUGAAAUUAAUUAAAUCACGC .((((((((((.....((((...((((((((..((((......))))......)))))))).-.))))..)))))))))).......((((((((((............)))))))).)) ( -35.20) >DroYak_CAF1 87780 119 - 1 ACUCCUGAAGGAGUUCAACUUUUGGCCUGAUAUUCCCUGAAUUGGGACUUCAAGUCGGGUUU-CAGUUGGCUUUCAAGGGGAAUUUUGGUGAUUUAAAAUUGAAAUUAAUUAAAUCAAGC .((((((((((...((((((...((((((((..((((......))))......)))))))).-.))))))))))).)))))(((((..((........))..)))))............. ( -35.00) >consensus ACUCCUGAAGGACUUAAACUUUUGGCCUGAUAUUCCCCGAAUUGGGACUUCGAGUCGGGUUU_GAGUUGGCUUUCAGGGGGAAUUUAGCUGAUUUAAAAUUGAAAUUAAUUAAAUCAAGC .((((((((((.....(((((..((((((((..((((......))))......))))))))..)))))..)))))))))).......((((((((((............)))))))).)) (-33.06 = -34.26 + 1.20)


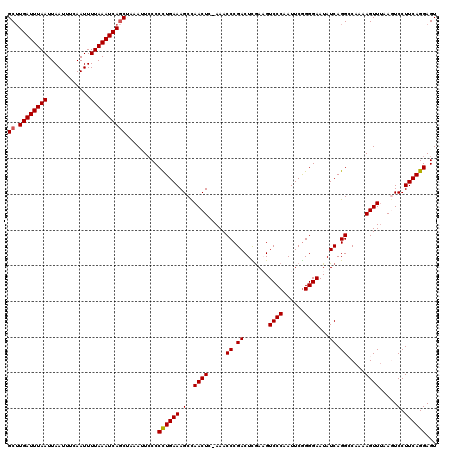
| Location | 11,350,306 – 11,350,408 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -29.09 |
| Consensus MFE | -19.40 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11350306 102 + 27905053 CCCCUGAAAGCCAACUCAAAACCCGACUCGAAGUCCCAAUUCGGGGAAUAUCAGGCCAAAAGUUUAAGUCCUUCAGGAGUUGCGCGAUUCGCAUUCAGGCAA------------------ ..((((((.((.(((((.......((((.((..((((......))))...))((((.....)))).))))......)))))))(((...))).))))))...------------------ ( -26.92) >DroSec_CAF1 92542 102 + 1 CCCCUGAAAGCUAACUCCAAACCCGACUCAACGUCCCAAUUCGGGGAAUAUCAGGGCAAAAGUUUAAGUCCUUCAGGAGUUGCACGAUUCGCAUUCAGGCAA------------------ ..((((((.(((((((((...(((((..............))))).......(((((..........)))))...)))))))........)).))))))...------------------ ( -28.64) >DroSim_CAF1 87731 120 + 1 CCCCUGAAAGCCAACUCAAAACCCGACUCGACGUCCCAAUUCGGGGAAUAUCAGGGCAAAAGUUUAAGUCCUUCAGGAGUUGCACGAUUCGCAUUCAGGCAAGCCAUGGCGAAUUUCCCU .((((((................((......))((((......))))...)))))).....((.(((.(((....))).))).))(((((((.....((....))...)))))))..... ( -30.20) >DroEre_CAF1 85521 101 + 1 CCCCUGAAAGCCAACUG-AAACCCGACUUGAAGUCCCAAUUCGGGGAAUAUCAGGCCAAAAGUUUAACUCCUUCAGGAGUUGCGCGAUUCGCAUUCAGACAA------------------ ...(((((.((...(((-(..(((((.(((......))).))))).....)))).......((.(((((((....))))))).)).....)).)))))....------------------ ( -32.30) >DroYak_CAF1 87820 97 + 1 CCCUUGAAAGCCAACUG-AAACCCGACUUGAAGUCCCAAUUCAGGGAAUAUCAGGCCAAAAGUUGAACUCCUUCAGGAGUUGCGCGAUUCGCAUUC----AA------------------ ....((((.(((((((.-......(.(((((..((((......))))...))))))....)))))((((((....))))))))(((...))).)))----).------------------ ( -27.40) >consensus CCCCUGAAAGCCAACUC_AAACCCGACUCGAAGUCCCAAUUCGGGGAAUAUCAGGCCAAAAGUUUAAGUCCUUCAGGAGUUGCGCGAUUCGCAUUCAGGCAA__________________ ..((((((.((..........((.((.......((((......))))...)).))......((.(((.(((....))).))).)).....)).))))))..................... (-19.40 = -20.16 + 0.76)

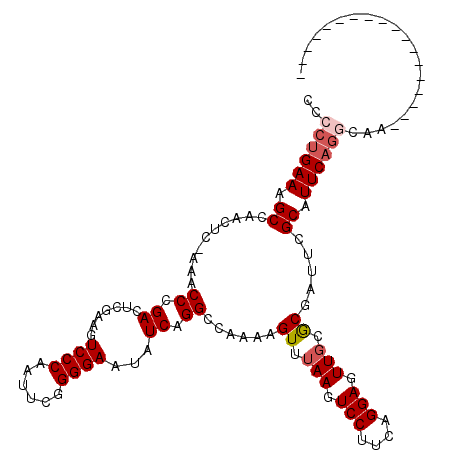
| Location | 11,350,306 – 11,350,408 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -31.86 |
| Energy contribution | -32.62 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11350306 102 - 27905053 ------------------UUGCCUGAAUGCGAAUCGCGCAACUCCUGAAGGACUUAAACUUUUGGCCUGAUAUUCCCCGAAUUGGGACUUCGAGUCGGGUUUUGAGUUGGCUUUCAGGGG ------------------.........((((.....)))).((((((((((.((..(((((..((((((((..((((......))))......))))))))..))))))))))))))))) ( -37.40) >DroSec_CAF1 92542 102 - 1 ------------------UUGCCUGAAUGCGAAUCGUGCAACUCCUGAAGGACUUAAACUUUUGCCCUGAUAUUCCCCGAAUUGGGACGUUGAGUCGGGUUUGGAGUUAGCUUUCAGGGG ------------------((((.(((.......))).))))((((((((((.((..((((((.((((.(((..((((......))))......)))))))..)))))))))))))))))) ( -33.50) >DroSim_CAF1 87731 120 - 1 AGGGAAAUUCGCCAUGGCUUGCCUGAAUGCGAAUCGUGCAACUCCUGAAGGACUUAAACUUUUGCCCUGAUAUUCCCCGAAUUGGGACGUCGAGUCGGGUUUUGAGUUGGCUUUCAGGGG .(.(..((((((...((....)).....))))))..).)..((((((((((.((..(((((..((((((((..((((......)))).))))....))))...))))))))))))))))) ( -40.10) >DroEre_CAF1 85521 101 - 1 ------------------UUGUCUGAAUGCGAAUCGCGCAACUCCUGAAGGAGUUAAACUUUUGGCCUGAUAUUCCCCGAAUUGGGACUUCAAGUCGGGUUU-CAGUUGGCUUUCAGGGG ------------------.........((((.....)))).((((((((((.....((((...((((((((..((((......))))......)))))))).-.))))..)))))))))) ( -34.10) >DroYak_CAF1 87820 97 - 1 ------------------UU----GAAUGCGAAUCGCGCAACUCCUGAAGGAGUUCAACUUUUGGCCUGAUAUUCCCUGAAUUGGGACUUCAAGUCGGGUUU-CAGUUGGCUUUCAAGGG ------------------((----(((.(((...)))((((((((....))))))(((((...((((((((..((((......))))......)))))))).-.))))))).)))))... ( -32.20) >consensus __________________UUGCCUGAAUGCGAAUCGCGCAACUCCUGAAGGACUUAAACUUUUGGCCUGAUAUUCCCCGAAUUGGGACUUCGAGUCGGGUUU_GAGUUGGCUUUCAGGGG ...........................((((.....)))).((((((((((.....(((((..((((((((..((((......))))......))))))))..)))))..)))))))))) (-31.86 = -32.62 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:30 2006