
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,349,254 – 11,349,494 |
| Length | 240 |
| Max. P | 0.999624 |

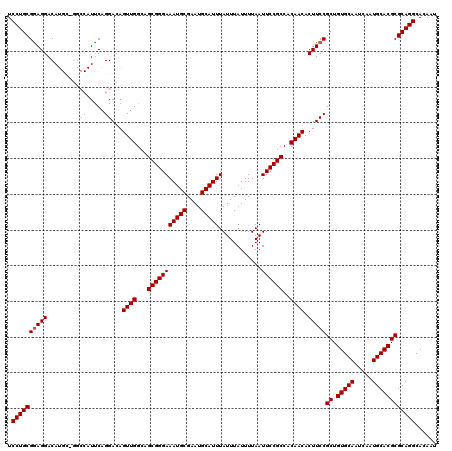
| Location | 11,349,254 – 11,349,374 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -39.44 |
| Consensus MFE | -35.12 |
| Energy contribution | -36.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11349254 120 + 27905053 UCCUGCGAAGGACAUGCAGGCCAUUCAGGACAGUUGGCAGCGGGAAAUGCGAAUGCAUUUAUUUAUUUUAAUUCCGCCACAACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAU ..(((((.......)))))(((.....(((..((((...(((((((((((....)))))............))))))..))))...)))((((((((.....)))))).)).)))..... ( -37.20) >DroSec_CAF1 88124 120 + 1 UCCUGCGGAGGACAUGCCGGCCAUUCAGGACAGUUGGCAGCGGGAAAUGCGAAUGCAUUUAUUUAUUUUAAUUCCGCCACAACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAU .((((((((((...(((((((...........)))))))(((((((((((....)))))............)))))).......)))))((.(((((.....))))))))))))...... ( -42.70) >DroSim_CAF1 86708 120 + 1 UCCUGCGGAGGACAUGCCGGCCAUUCAGGACAGUUGGCAGCGGGAAAUGCGAAUGCAUUUAUUUAUUUUAAUUCCGCCACAACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAU .((((((((((...(((((((...........)))))))(((((((((((....)))))............)))))).......)))))((.(((((.....))))))))))))...... ( -42.70) >DroEre_CAF1 84373 111 + 1 UCCUGCGGAGGACA---------UCCAGGACAGUUGGCAGCGGGAAAUGCGAAUGCAUUUAUUUAUUUUAAUUCCGCCACAACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAU .((((((((.....---------))).(((..((((...(((((((((((....)))))............))))))..))))...)))((.(((((.....))))))))))))...... ( -37.50) >DroYak_CAF1 86671 111 + 1 UCCUGCGGAGGACA---------UUCAGGACAGUUGGCAGCGGGAAAUGCGAAUGCAUUUAUUUAUUUUAAUUCCGCCACAACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAU .((((((((((.(.---------....)....((((...(((((((((((....)))))............))))))..)))).)))))((.(((((.....))))))))))))...... ( -37.10) >consensus UCCUGCGGAGGACAUGC_GGCCAUUCAGGACAGUUGGCAGCGGGAAAUGCGAAUGCAUUUAUUUAUUUUAAUUCCGCCACAACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAU .((((((((((.........((.....))...((((...(((((((((((....)))))............))))))..)))).)))))((.(((((.....))))))))))))...... (-35.12 = -36.12 + 1.00)


| Location | 11,349,254 – 11,349,374 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.63 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -38.30 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.80 |
| SVM RNA-class probability | 0.999624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11349254 120 - 27905053 AUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCGCUGCCAACUGUCCUGAAUGGCCUGCAUGUCCUUCGCAGGA ......((((((((((((.....))))).))(((...((((..((((.(((......)))((((((....)))))).))))..).)))..))).(((.(((......))).)))))))). ( -41.20) >DroSec_CAF1 88124 120 - 1 AUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCGCUGCCAACUGUCCUGAAUGGCCGGCAUGUCCUCCGCAGGA ......((((((((((((.....))))).))(((.((((((.(((((.(((......)))((((((....)))))).)))))((((...........)))))))))).)))...))))). ( -42.00) >DroSim_CAF1 86708 120 - 1 AUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCGCUGCCAACUGUCCUGAAUGGCCGGCAUGUCCUCCGCAGGA ......((((((((((((.....))))).))(((.((((((.(((((.(((......)))((((((....)))))).)))))((((...........)))))))))).)))...))))). ( -42.00) >DroEre_CAF1 84373 111 - 1 AUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCGCUGCCAACUGUCCUGGA---------UGUCCUCCGCAGGA ......((((((((((((.....))))).))(((...((((..((((.(((......)))((((((....)))))).))))..).)))..))).(((---------.....)))))))). ( -40.00) >DroYak_CAF1 86671 111 - 1 AUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCGCUGCCAACUGUCCUGAA---------UGUCCUCCGCAGGA ......((((((((((((.....))))).))(((...((((..((((.(((......)))((((((....)))))).))))..).)))..)))....---------........))))). ( -38.30) >consensus AUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCGCUGCCAACUGUCCUGAAUGGCC_GCAUGUCCUCCGCAGGA ......((((((((((((.....))))).))(((...((((..((((.(((......)))((((((....)))))).))))..).)))..))).....................))))). (-38.30 = -38.30 + 0.00)

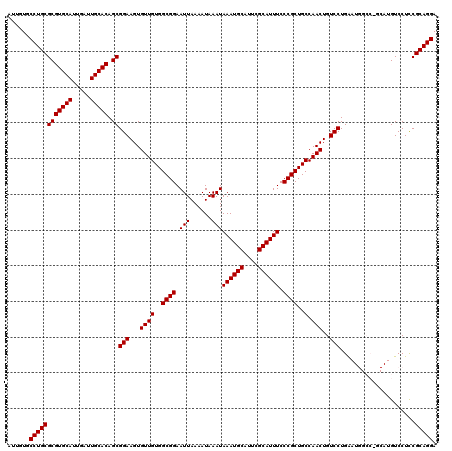
| Location | 11,349,294 – 11,349,414 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -31.28 |
| Energy contribution | -31.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11349294 120 - 27905053 UGGAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCG .(((((((((....(((((....)))))..))))))))).(((.((((.((..(((((.....))))).((....))))...)))).)))..........((((((....)))))).... ( -31.90) >DroSec_CAF1 88164 118 - 1 --GAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCG --((((((((....(((((....)))))..))))))))..(((.((((.((..(((((.....))))).((....))))...)))).)))..........((((((....)))))).... ( -31.20) >DroSim_CAF1 86748 118 - 1 --GAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCG --((((((((....(((((....)))))..))))))))..(((.((((.((..(((((.....))))).((....))))...)))).)))..........((((((....)))))).... ( -31.20) >DroEre_CAF1 84404 118 - 1 --GAUAUGUAUUUACAUGUAUGCAUAUGGCUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCG --.((((((((........))))))))(((.(((........)))))).((..(((((.....))))).))((((((((....(((..(((......)))...)))....)))))))).. ( -31.60) >DroYak_CAF1 86702 118 - 1 --GAUAUGUAUUCACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCG --((((((((..((.(((....))).))..))))))))..(((.((((.((..(((((.....))))).((....))))...)))).)))..........((((((....)))))).... ( -32.90) >consensus __GAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUUGUGGCGGAAUUAAAAUAAAUAAAUGCAUUCGCAUUUCCCG ..((((((((.(((.(((....))).))).))))))))..(((.((((.((..(((((.....))))).((....))))...)))).)))..........((((((....)))))).... (-31.28 = -31.12 + -0.16)


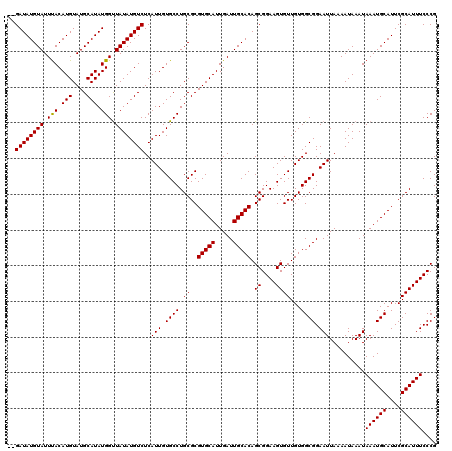
| Location | 11,349,334 – 11,349,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11349334 120 + 27905053 AACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAUGAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUCCAUCCAUUUAUGGAGGCAGCUCUUCAUACAUAUAUGUACUAU ....(((.(((.(((((.....)))))))).))).......(((((((((...(((((....)))))...................((((((((....))))))))..))))))).)).. ( -25.10) >DroSec_CAF1 88204 105 + 1 AACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAUGAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUACA-----------U ....(((.(((.(((((.....)))))))).)))...((((.(((((((....))))).....((((....)))).))----))))((((((((....))))))))..-----------. ( -20.70) >DroSim_CAF1 86788 105 + 1 AACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAUGAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUACA-----------U ....(((.(((.(((((.....)))))))).)))...((((.(((((((....))))).....((((....)))).))----))))((((((((....))))))))..-----------. ( -20.70) >DroEre_CAF1 84444 105 + 1 AACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAUGAGACAUAUAGCCAUAUGCAUACAUGUAAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUACG-----------C ....(((.(((.(((((.....)))))))).)))....(((((.......((((((((....))))).........((----((....))))))).....)))))...-----------. ( -22.40) >DroYak_CAF1 86742 102 + 1 AACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAUGAGACAUAUAACCAUAUGCAUACAUGUGAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUA-------------- ....(((.(((.(((((.....)))))))).)))...((((.((.((.((..((((((....))))))..)).)).))----))))((((((((....))))))))-------------- ( -22.90) >consensus AACACUUCCGCUGUGCAAUCAAUGCACGCGCAGGCACAAUGAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUC____CAUUUAUGGAGGCAGCUCUUCAUACA___________U ....(((.(((.(((((.....)))))))).)))...((((.(.(((((....))))))....((((....)))).......))))((((((((....)))))))).............. (-20.10 = -20.10 + -0.00)

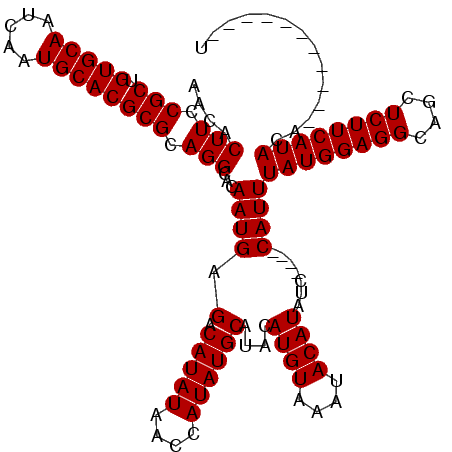
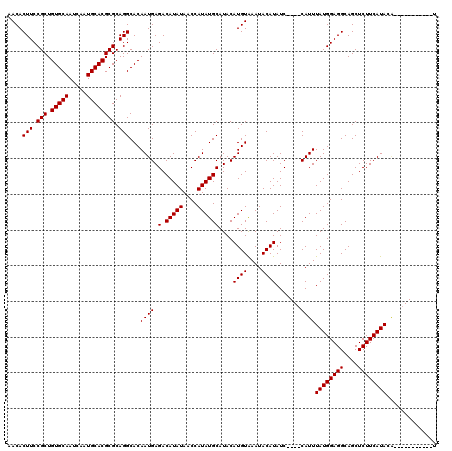
| Location | 11,349,334 – 11,349,454 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11349334 120 - 27905053 AUAGUACAUAUAUGUAUGAAGAGCUGCCUCCAUAAAUGGAUGGAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUU ...(((((((((((((..........((((((....)))).)).........)))))))))((((((....))))))......))))((((..(((((.....))))).))))....... ( -32.81) >DroSec_CAF1 88204 105 - 1 A-----------UGUAUGAAGAGCUGCCUCCAUAAAUG----GAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUU .-----------.(((..(.((((.(((((((....))----))(((((((........))))))).))).....).))).)..)))((((..(((((.....))))).))))....... ( -29.30) >DroSim_CAF1 86788 105 - 1 A-----------UGUAUGAAGAGCUGCCUCCAUAAAUG----GAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUU .-----------.(((..(.((((.(((((((....))----))(((((((........))))))).))).....).))).)..)))((((..(((((.....))))).))))....... ( -29.30) >DroEre_CAF1 84444 105 - 1 G-----------CGUAUGAAGAGCUGCCUCCAUAAAUG----GAUAUGUAUUUACAUGUAUGCAUAUGGCUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUU .-----------.(((..(.((((.(((((((....))----))(((((((........))))))).))).....).))).)..)))((((..(((((.....))))).))))....... ( -31.20) >DroYak_CAF1 86742 102 - 1 --------------UAUGAAGAGCUGCCUCCAUAAAUG----GAUAUGUAUUCACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUU --------------........((.((...(((((..(----((((((((..((.(((....))).))..)))))))))..)))))...))))(((((.....))))).((....))... ( -29.30) >consensus A___________UGUAUGAAGAGCUGCCUCCAUAAAUG____GAUAUGUAUUUACAUGUAUGCAUAUGGUUAUAUGUCUCAUUGUGCCUGCGCGUGCAUUGAUUGCACAGCGGAAGUGUU .............(((..(.((((.....(((((..((.....((((((....))))))...)))))))......).))).)..)))((((..(((((.....))))).))))....... (-25.92 = -26.12 + 0.20)


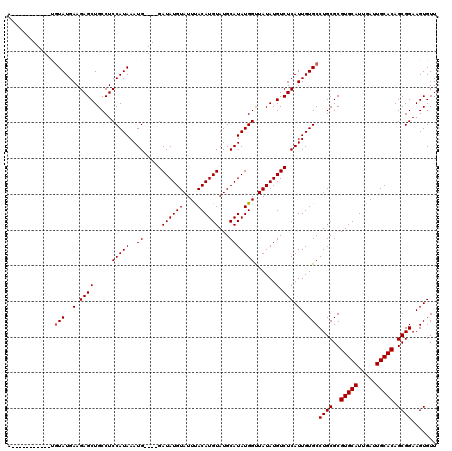
| Location | 11,349,374 – 11,349,494 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -19.08 |
| Energy contribution | -20.70 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11349374 120 + 27905053 GAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUCCAUCCAUUUAUGGAGGCAGCUCUUCAUACAUAUAUGUACUAUGUGCGCUUGUGUGUGUACUUAUGCAUGUCCAGUGUCCUGU ..(((((......(((((((((...(((.((((((((((((......)))))(((.((.(....((((....))))....).)))))))))))).))).)))))))))...))))).... ( -31.00) >DroSec_CAF1 88244 105 + 1 GAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUACA-----------UGUGCGCUUGUGUGUGUACUUAUGCAUGUCCUGUGUCCCCU ..((((((.....(((((((((...(((.(((((((.(----(((.((((((((....)))))))).)-----------)).)....))))))).))).)))))))))..)))))).... ( -29.90) >DroSim_CAF1 86828 100 + 1 GAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUACA-----------UGUGCGCUUGUGUGUGAACUUAUGCAUGUCCUGUGU----- ...(((((.....(((((((((...((..(((((((.(----(((.((((((((....)))))))).)-----------)).)....)))))))..)).)))))))))..)))))----- ( -25.10) >DroEre_CAF1 84484 97 + 1 GAGACAUAUAGCCAUAUGCAUACAUGUAAAUACAUAUC----CAUUUAUGGAGGCAGCUCUUCAUACG-----------CG--------UAUAUGUAGGUGUGCAUGUCCUGCGUCCUGC ..........((...(((((.(((((((..(((((((.----(...((((((((....))))))))..-----------.)--------.)))))))....)))))))..)))))...)) ( -25.10) >consensus GAGACAUAUAACCAUAUGCAUACAUGUAAAUACAUAUC____CAUUUAUGGAGGCAGCUCUUCAUACA___________UGUGCGCUUGUGUGUGUACUUAUGCAUGUCCUGUGUCCUGU ..((((((.....(((((((((...(((.(((((((..........((((((((....))))))))((((.......))))......))))))).))).)))))))))..)))))).... (-19.08 = -20.70 + 1.62)

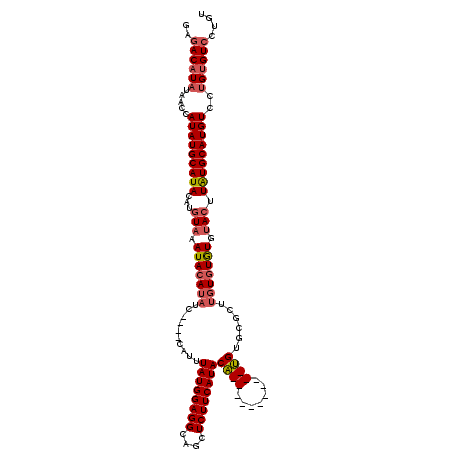

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:23 2006