
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,345,333 – 11,345,558 |
| Length | 225 |
| Max. P | 0.988230 |

| Location | 11,345,333 – 11,345,450 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.94 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.46 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11345333 117 + 27905053 CUUUAUUGCAGUUUCAGCAGCAGCUCCUGCAUUGUCCCUGCUGUUAUCGCUGCUUUUCAAAUGACAUUUAAAUAUUUUCCACAAAGCCACAAGCUGUCAGGGAAUUUAGAAUAGUGG--- .......(((((...((((((((...(......)...))))))))...)))))...........((((....((..((((....(((.....))).....))))..))....)))).--- ( -28.20) >DroSec_CAF1 84197 117 + 1 CUUUAUUGCAGUUUCAGCAGCAGCUCCUGCAUUGUCCCUGCUGUUAUCUCUGCUUUUCAAAUGACAUUUAAAUAUUUCCCACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG--- .......((((....((((((((...(......)...))))))))....))))...........((((....((((((((....(((.....)))....))))).)))....)))).--- ( -28.10) >DroSim_CAF1 82743 117 + 1 CUUUAUUGCAGUUUCAGCAGCAGCUCCUGCAUUGUCCCUGCUGUUAUCGCUGCUUUUCAAAUGACAUUUAAAUAUUUCCCACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG--- .......(((((...((((((((...(......)...))))))))...)))))...........((((....((((((((....(((.....)))....))))).)))....)))).--- ( -29.50) >DroEre_CAF1 80477 120 + 1 CUUUAUUGCAGUUUCAGCAGCAGCUUCUGCAUUGUCCCUGCUGUUAUCGCAGCUUUUCAAACGACAUUUAAAUAUUUCCCACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGGGGGAC ......(((((....(((....))).)))))..(((((((((((....)))))............((((......(((((....(((.....)))....)))))....))))..)))))) ( -32.80) >DroYak_CAF1 82590 117 + 1 CUUUAUUGCAGUUUCAGCAGCAGCUCCGGCAUUGUCCCUGCUGUUAUCGCUGCUUUUCAAAUGACAUUUAAAUAUUUCCCACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG--- .......(((((...((((((((....(((...))).))))))))...)))))...........((((....((((((((....(((.....)))....))))).)))....)))).--- ( -29.90) >consensus CUUUAUUGCAGUUUCAGCAGCAGCUCCUGCAUUGUCCCUGCUGUUAUCGCUGCUUUUCAAAUGACAUUUAAAUAUUUCCCACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG___ .......(((((...((((((((...(......)...))))))))...)))))...........((((....((((((((....(((.....)))....))))).)))....)))).... (-25.98 = -26.46 + 0.48)



| Location | 11,345,413 – 11,345,519 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -26.59 |
| Energy contribution | -27.23 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11345413 106 + 27905053 ACAAAGCCACAAGCUGUCAGGGAAUUUAGAAUAGUGG------CAGUAUGCCCAGGAC--------UAAGGACACUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCA ............(((((((((..(((......)))((------((((((..((.....--------...))((((((((......))))).)))....))))))))..)))))))))... ( -33.90) >DroSec_CAF1 84277 106 + 1 ACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG------GAAUAUGGCCAGGAC--------UAAGGACGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCA .....((((((....(((..((....(((.(((....------...)))..)))...)--------)...)))((((((......)))))))))).....((((((.....)))))))). ( -32.70) >DroSim_CAF1 82823 106 + 1 ACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG------GAAUACGGCCAGGAC--------UAAGGACGCUCCUGCCACAAGGAGCUGUAAAGAUGCUGUCGGUCUGACAGCGCA ............(((((((((.....(((....(((.------...)))..)))....--------....(((((((((......)))))).(((....))).)))..)))))))))... ( -33.00) >DroEre_CAF1 80557 120 + 1 ACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGGGGGACCAGGAAUAUGGCCAGGACUCCUGGCCUAAGGAUGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCA .....((((((....(((...............((....))........(((((((...)))))))....)))((((((......)))))))))).....((((((.....)))))))). ( -44.60) >DroYak_CAF1 82670 106 + 1 ACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG------GAAUAUGGCCAGGAC--------CAAGGAUGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGCUCUGACAGCGCA ............((((((((((...((((.(((((..------.....((((.((((.--------((....)).))))))))......))))).......))))..))))))))))... ( -33.42) >consensus ACAAAGCCACAAGCUGUCAGGGAAUAUGGAAUAGUGG______GAAUAUGGCCAGGAC________UAAGGACGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCA .....((((((....(((..((.((((..................))))..))..)))...............((((((......)))))))))).....((((((.....)))))))). (-26.59 = -27.23 + 0.64)



| Location | 11,345,413 – 11,345,519 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -27.07 |
| Energy contribution | -26.99 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11345413 106 - 27905053 UGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGUGUCCUUA--------GUCCUGGGCAUACUG------CCACUAUUCUAAAUUCCCUGACAGCUUGUGGCUUUGU .((((((((.....)))))).....(((((((....(((((((..((((((...--------.....)))))).)))------))))...((..........)).))).))))))..... ( -38.60) >DroSec_CAF1 84277 106 - 1 UGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCGUCCUUA--------GUCCUGGCCAUAUUC------CCACUAUUCCAUAUUCCCUGACAGCUUGUGGCUUUGU .((((((((.....)))))).....(((((((...((((((((((.(.......--------))))).))))))...------.........((.......))..))).))))))..... ( -28.90) >DroSim_CAF1 82823 106 - 1 UGCGCUGUCAGACCGACAGCAUCUUUACAGCUCCUUGUGGCAGGAGCGUCCUUA--------GUCCUGGCCGUAUUC------CCACUAUUCCAUAUUCCCUGACAGCUUGUGGCUUUGU .((((((((.....)))))).....((((((((((......)))))).......--------(((..((..((((..------..........))))..)).)))....))))))..... ( -27.20) >DroEre_CAF1 80557 120 - 1 UGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCAUCCUUAGGCCAGGAGUCCUGGCCAUAUUCCUGGUCCCCCUAUUCCAUAUUCCCUGACAGCUUGUGGCUUUGU .((((((((.....)))))).....((((((((((......)))))).......(((((((...))))))).......(((..........)))...............))))))..... ( -39.30) >DroYak_CAF1 82670 106 - 1 UGCGCUGUCAGAGCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCAUCCUUG--------GUCCUGGCCAUAUUC------CCACUAUUCCAUAUUCCCUGACAGCUUGUGGCUUUGU ...((((((((.((....))...............((((((((((.((....))--------.)))).))))))...------.................))))))))............ ( -32.80) >consensus UGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCGUCCUUA________GUCCUGGCCAUAUUC______CCACUAUUCCAUAUUCCCUGACAGCUUGUGGCUUUGU .((((((((.....)))))).....((((((((((......))))))...............(((..((..((((..................))))..)).)))....))))))..... (-27.07 = -26.99 + -0.08)



| Location | 11,345,450 – 11,345,558 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -42.46 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.96 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11345450 108 + 27905053 ---CAGUAUGCCCAGGAC--------UAAGGACACUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCACAAAGCUCAAAUUAGUGGCGGCCUCGCCCUCGGCCGUCG- ---............(((--------..(((.....)))(((((((.((((((((....(((((((.....))))))))))..)))))...)).)))))((((........))))))).- ( -39.70) >DroSec_CAF1 84314 108 + 1 ---GAAUAUGGCCAGGAC--------UAAGGACGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCACAAAGCUCAAAUUAGUGGCAACCUCGCCCUCGGCCGUCG- ---....(((((((((..--------..(((.....)))(((((((.((((((((....(((((((.....))))))))))..)))))...)).)))))........))).))))))..- ( -42.20) >DroSim_CAF1 82860 108 + 1 ---GAAUACGGCCAGGAC--------UAAGGACGCUCCUGCCACAAGGAGCUGUAAAGAUGCUGUCGGUCUGACAGCGCACAAAGCUCAAAUUAGUGGCAACCUCGCCCUCGGCCGUCG- ---....(((((((((..--------..(((.....)))(((((((.(((((.....(.(((((((.....))))))))....)))))...)).)))))........))).))))))..- ( -43.30) >DroEre_CAF1 80597 119 + 1 CAGGAAUAUGGCCAGGACUCCUGGCCUAAGGAUGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCACAAAGCUCAAAUUAGUGGUGGCCUCGCCCUCG-CCCUCGG .(((.....(((((((...)))))))..(((..((((((......)))))).((((....((((((.....))))))((.((..(((......)))..)))).)))))))..-.)))... ( -44.80) >DroYak_CAF1 82707 109 + 1 ---GAAUAUGGCCAGGAC--------CAAGGAUGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGCUCUGACAGCGCACAAAGCUCAAAUUAGUGGCGGCCUCGCCCUCGGCACUCGG ---......((((((((.--------((....)).))))(((((((.((((((((....(((((((.....))))))))))..)))))...)).)))))))))..(((...)))...... ( -42.30) >consensus ___GAAUAUGGCCAGGAC________UAAGGACGCUCCUGCCACAAGGAGCUGUGAAGAUGCUGUCGGUCUGACAGCGCACAAAGCUCAAAUUAGUGGCGGCCUCGCCCUCGGCCGUCG_ .......(((((((((............(((.....)))(((((((.(((((.....(.(((((((.....))))))))....)))))...)).)))))........))).))))))... (-36.20 = -36.96 + 0.76)

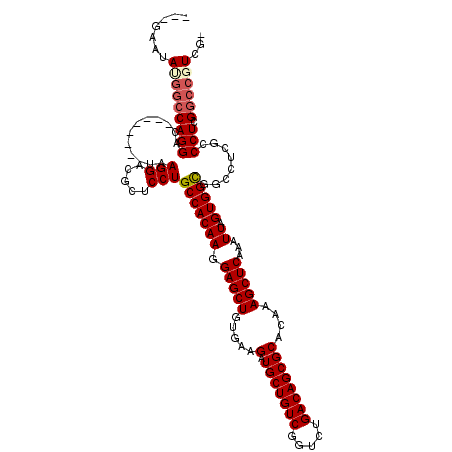

| Location | 11,345,450 – 11,345,558 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -46.24 |
| Consensus MFE | -38.14 |
| Energy contribution | -38.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11345450 108 - 27905053 -CGACGGCCGAGGGCGAGGCCGCCACUAAUUUGAGCUUUGUGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGUGUCCUUA--------GUCCUGGGCAUACUG--- -.....(((.((((((((((((((((......(((((.((...((((((.....)))))).....)).)))))...))))).)).....)))).--------))))).)))......--- ( -44.70) >DroSec_CAF1 84314 108 - 1 -CGACGGCCGAGGGCGAGGUUGCCACUAAUUUGAGCUUUGUGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCGUCCUUA--------GUCCUGGCCAUAUUC--- -....((((.((((((((((((((((......(((((.((...((((((.....)))))).....)).)))))...)))))))......)))).--------)))))))))......--- ( -45.50) >DroSim_CAF1 82860 108 - 1 -CGACGGCCGAGGGCGAGGUUGCCACUAAUUUGAGCUUUGUGCGCUGUCAGACCGACAGCAUCUUUACAGCUCCUUGUGGCAGGAGCGUCCUUA--------GUCCUGGCCGUAUUC--- -..((((((.((((((((((((((((......(((((.((...((((((.....)))))).....)).)))))...)))))))......)))).--------)))))))))))....--- ( -46.90) >DroEre_CAF1 80597 119 - 1 CCGAGGG-CGAGGGCGAGGCCACCACUAAUUUGAGCUUUGUGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCAUCCUUAGGCCAGGAGUCCUGGCCAUAUUCCUG ..(((((-.(((((((((((((.........)).)))))))..((((((.....))))))..))))...((((((......)))))).))))).(((((((...)))))))......... ( -49.30) >DroYak_CAF1 82707 109 - 1 CCGAGUGCCGAGGGCGAGGCCGCCACUAAUUUGAGCUUUGUGCGCUGUCAGAGCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCAUCCUUG--------GUCCUGGCCAUAUUC--- ..(((((((...)))..(((((((((......(((((.((...((((((.....)))))).....)).)))))...)))))((((.((....))--------.))))))))..))))--- ( -44.80) >consensus _CGACGGCCGAGGGCGAGGCCGCCACUAAUUUGAGCUUUGUGCGCUGUCAGACCGACAGCAUCUUCACAGCUCCUUGUGGCAGGAGCGUCCUUA________GUCCUGGCCAUAUUC___ .....((((.(((((......(((((......(((((.((...((((((.....)))))).....)).)))))...)))))(((.....)))..........)))))))))......... (-38.14 = -38.78 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:12 2006