| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,345,173 – 11,345,306 |
| Length | 133 |
| Max. P | 0.794265 |

| Location | 11,345,173 – 11,345,280 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.96 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -21.15 |
| Energy contribution | -22.23 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
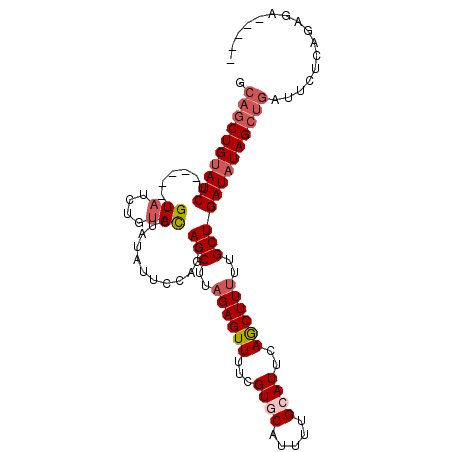
>3R_DroMel_CAF1 11345173 107 - 27905053 ACCUAUAUUCCUGCAGCUUAGAGUUUUCGUGCAUUUUGCAUUCAACUUUUUGCUGAUAUAGCUGAUUCUCAGAGA-------------AUUGCGGUUUGCUGGGAUUUAGUUAGAGGUCU ((((.........((((..((((((...((((.....))))..))))))..))))...(((((((.((((((..(-------------((....)))..)))))).))))))).)))).. ( -34.30) >DroSec_CAF1 84040 107 - 1 ACCUAUAUUCCAGCAGCUUAGAGUUUUCGUUCAUUUUGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCUCUGAGA-------------AUUGCGGUUUGCUGGGAUUUAGUUAGAGGUCU ..(((.(((((((((((((((((...((((.......))..(((((.....))))).......))..))))))).-------------........)))))))))).))).......... ( -30.00) >DroSim_CAF1 82587 106 - 1 ACCUAUAUUCCAGCAGCUUAGAGUUUUCGUUCAUUUUGCAUUCAGCUU-UUGCUGAUAUAGCUGAUUCUCAGAGA-------------AUUGCGGUUUGCUGGGAUUUAGUUAGAGGUCU ..(((.((((((((((((...(((((((.........((..(((((..-..)))))....)).........))))-------------)))..)).)))))))))).))).......... ( -30.07) >DroEre_CAF1 80312 120 - 1 AUGUAUAUCCCUGCAGCUCAGAGUUUUCGUGCAUUUGGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCCCGGAGAAGGAAGACGUUGAAGUGCGGUUUGCUGGCAUUUAUUUAGAGGUUU ((((......(((((..((((.((((((.(...(((((...((((((............))))))...)))))..).)))))).))))..))))).......)))).............. ( -31.12) >DroYak_CAF1 82399 120 - 1 ACCUAUAUCCCUGCAGCUCAGAGUUUUCGUGCAUUUUGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCCCGCAGAAGGGAGACGUUGAAGUGCGGUUUGCUGGGAUUUAUUUAGAGGUUU ((((..(((((.((((..(.(......)..(((((((((..(((((.....)))))....))...(((((......))))).....))))))))..)))).)))))........)))).. ( -36.00) >consensus ACCUAUAUUCCUGCAGCUUAGAGUUUUCGUGCAUUUUGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCUCAGAGA_____________AUUGCGGUUUGCUGGGAUUUAGUUAGAGGUCU ((((.........((((..((((((...((((.....))))..))))))..))))...(((((((.((((((...........................)))))).))))))).)))).. (-21.15 = -22.23 + 1.08)


| Location | 11,345,205 – 11,345,306 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.58 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -16.70 |
| Energy contribution | -18.32 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11345205 101 - 27905053 UCAGCUGUAUCUGUAUCUGUAUCUGUACCUAUAUUCCUGCAGCUUAGAGUUUUCGUGCAUUUUGCAUUCAACUUUUUGCUGAUAUAGCUGAUUCUCAGAGA----- (((((((((((.(((...((((........))))...)))(((..((((((...((((.....))))..))))))..))))))))))))))..........----- ( -28.90) >DroSec_CAF1 84072 95 - 1 GCAGCUGUAUCU------GUAUCUGUACCUAUAUUCCAGCAGCUUAGAGUUUUCGUUCAUUUUGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCUCUGAGA----- .(((((((((((------(...((((............))))..))))...................(((((.....)))))))))))))...........----- ( -21.50) >DroSim_CAF1 82619 94 - 1 GCAGCUGUAUCU------GUAUCUGUACCUAUAUUCCAGCAGCUUAGAGUUUUCGUUCAUUUUGCAUUCAGCUU-UUGCUGAUAUAGCUGAUUCUCAGAGA----- .(((((((((((------(...((((............))))..))))...................(((((..-..)))))))))))))...........----- ( -21.50) >DroEre_CAF1 80352 94 - 1 ----CUGCAUCC------GUACCU--AUGUAUAUCCCUGCAGCUCAGAGUUUUCGUGCAUUUGGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCCCGGAGAAGGAA ----.....(((------((((..--..)))).(((...(((((..........((((.....))))(((((.....)))))...)))))......)))...))). ( -21.90) >consensus GCAGCUGUAUCU______GUAUCUGUACCUAUAUUCCAGCAGCUUAGAGUUUUCGUGCAUUUUGCAUUCAGCUUUUUGCUGAUAUAGCUGAUUCUCAGAGA_____ .((((((((((.......(((....)))............(((..((((((...((((.....))))..))))))..)))))))))))))................ (-16.70 = -18.32 + 1.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:08 2006