
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,344,845 – 11,344,956 |
| Length | 111 |
| Max. P | 0.935421 |

| Location | 11,344,845 – 11,344,956 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.58 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
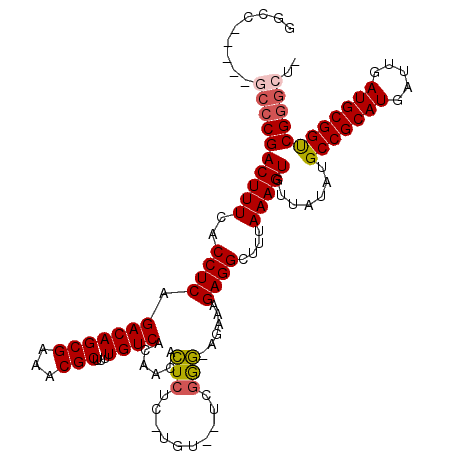
>3R_DroMel_CAF1 11344845 111 + 27905053 GGCC-----GCCCGACUUUCACCUCAGACAGCGAAACGCUUUUGUCACAACAUUCUA-UGU--UCGGAAAGAAAGAGGCUUUAAAGUGUUAUAUGCCGCAUGAUUGAUGCGGUCGGGCU- ....-----((((((((((..((((.(((((((...)))...))))......((((.-...--..)))).....))))....))))).......(((((((.....)))))))))))).- ( -34.20) >DroSec_CAF1 83714 110 + 1 GGCC-----GCCCGACUUUCACCUCAGACAGCGAAACGCUUUUGUCACAACACUCUC-UGU--UUGGG-AGAAAGAGGCUUUAAAGUGUUAUAUGCCGCAUGAUUGAUGCGGUCGGGCU- ....-----((((((((((..((((.(((((((...)))...))))......(((((-...--..)))-))...))))....))))).......(((((((.....)))))))))))).- ( -39.40) >DroSim_CAF1 82261 110 + 1 GGCC-----GCCCGACUUUCACCUCAGACAGCGAAACGCUUUUGUCACAACACUCUC-UGU--UUGGG-AGAAAGAGGCUUUAAAGUGUUAUAUGCCGCAUGAUUGAUGCGGUCGGGCU- ....-----((((((((((..((((.(((((((...)))...))))......(((((-...--..)))-))...))))....))))).......(((((((.....)))))))))))).- ( -39.40) >DroEre_CAF1 79985 116 + 1 GGCCGACUCGCCCGACUUUCACCUCAGACAGCGAUACGCUUUUGUCACAACACUCUC-UGC--UCGGG-AGAAAGAGGCUUUAAAGUGUUAUAUGCCGCAUGAUUGAUGCGGCCGGGGUU ..........(((((((((..((((.(((((((...)))...))))......(((((-...--..)))-))...))))....))))).......(((((((.....)))))))))))... ( -37.50) >DroYak_CAF1 82091 112 + 1 GCCCGA------CGACUUUCACCUCAGACAGCGAAACGCUUUUGUCACAACACUCUCCUUCUCACAAG-AGAAAGAGGCUUUAAAGUGUUAUAUGCCGCAUGAUUGAUGCGGUCGGGGU- .(((((------(.(((((..((((.(((((((...)))...))))............(((((....)-)))).))))....))))).........(((((.....)))))))))))..- ( -35.10) >consensus GGCC_____GCCCGACUUUCACCUCAGACAGCGAAACGCUUUUGUCACAACACUCUC_UGU__UCGGG_AGAAAGAGGCUUUAAAGUGUUAUAUGCCGCAUGAUUGAUGCGGUCGGGCU_ .........((((((((((..((((.(((((((...)))...))))......(((..........)))......))))....))))).......(((((((.....)))))))))))).. (-28.22 = -28.58 + 0.36)

| Location | 11,344,845 – 11,344,956 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.05 |
| Mean single sequence MFE | -38.54 |
| Consensus MFE | -29.39 |
| Energy contribution | -30.23 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344845 111 - 27905053 -AGCCCGACCGCAUCAAUCAUGCGGCAUAUAACACUUUAAAGCCUCUUUCUUUCCGA--ACA-UAGAAUGUUGUGACAAAAGCGUUUCGCUGUCUGAGGUGAAAGUCGGGC-----GGCC -.((((((((((((.....)))))).........((((...(((((..........(--(((-(...)))))..((((...(((...))))))).))))).))))))))))-----.... ( -35.00) >DroSec_CAF1 83714 110 - 1 -AGCCCGACCGCAUCAAUCAUGCGGCAUAUAACACUUUAAAGCCUCUUUCU-CCCAA--ACA-GAGAGUGUUGUGACAAAAGCGUUUCGCUGUCUGAGGUGAAAGUCGGGC-----GGCC -.((((((((((((.....))))))....................(((((.-((...--.((-((.((((....(((......))).)))).)))).)).)))))))))))-----.... ( -37.30) >DroSim_CAF1 82261 110 - 1 -AGCCCGACCGCAUCAAUCAUGCGGCAUAUAACACUUUAAAGCCUCUUUCU-CCCAA--ACA-GAGAGUGUUGUGACAAAAGCGUUUCGCUGUCUGAGGUGAAAGUCGGGC-----GGCC -.((((((((((((.....))))))....................(((((.-((...--.((-((.((((....(((......))).)))).)))).)).)))))))))))-----.... ( -37.30) >DroEre_CAF1 79985 116 - 1 AACCCCGGCCGCAUCAAUCAUGCGGCAUAUAACACUUUAAAGCCUCUUUCU-CCCGA--GCA-GAGAGUGUUGUGACAAAAGCGUAUCGCUGUCUGAGGUGAAAGUCGGGCGAGUCGGCC ....((((((((((.....)))))))...............(((((((((.-((...--.((-((.((((.(((.......)))...)))).)))).)).)))))..))))....))).. ( -40.00) >DroYak_CAF1 82091 112 - 1 -ACCCCGACCGCAUCAAUCAUGCGGCAUAUAACACUUUAAAGCCUCUUUCU-CUUGUGAGAAGGAGAGUGUUGUGACAAAAGCGUUUCGCUGUCUGAGGUGAAAGUCG------UCGGGC -..((((((.((((.....))))(((.(((((((((.......((((((((-(....)))))))))))))))))).........(((((((......)))))))))))------))))). ( -43.11) >consensus _AGCCCGACCGCAUCAAUCAUGCGGCAUAUAACACUUUAAAGCCUCUUUCU_CCCGA__ACA_GAGAGUGUUGUGACAAAAGCGUUUCGCUGUCUGAGGUGAAAGUCGGGC_____GGCC ..(((((((..((((..(((.(((((.(((((((((((..........................)))))))))))((......))...))))).)))))))...)))))))......... (-29.39 = -30.23 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:06 2006