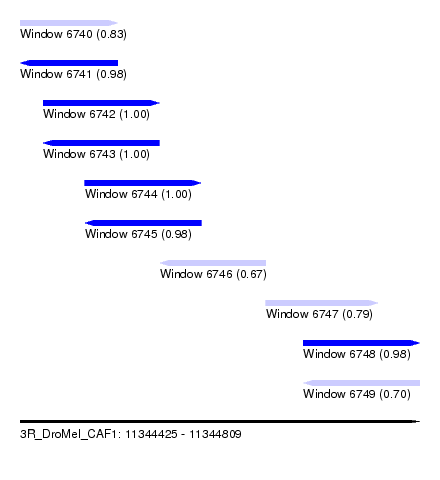
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,344,425 – 11,344,809 |
| Length | 384 |
| Max. P | 0.999693 |

| Location | 11,344,425 – 11,344,519 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -25.96 |
| Energy contribution | -25.52 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344425 94 + 27905053 UUU--GUGGGUGUGUGUGCCGUGGCAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCUGGCAGUGGCGAAGAAA-AAAGCCACCAGC ..(--((.(((....(((..(((.(((((((((((.....))))))))))))))..)))......))).)))(((((.......-...))))).... ( -28.80) >DroSec_CAF1 83293 95 + 1 UUU--GUGUGUGUGUGUGCCGUGGCAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCCGGCAGUGGCGGAGAAAAAAAGCCACCAGC ...--.....((.(((.((.(((.(((((((((((.....))))))))))))))....((((((.((((....)))).))))))....))))))).. ( -28.10) >DroSim_CAF1 81840 95 + 1 UUU--GUGUGUGUGUGUGCCGUGGCAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCCGGCAGUGGCGGAGAAAAAAAGCCACCAGC ...--.....((.(((.((.(((.(((((((((((.....))))))))))))))....((((((.((((....)))).))))))....))))))).. ( -28.10) >DroEre_CAF1 79547 95 + 1 UU-GGGUGUGUGUGUGCGCCGUGGCAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCUGGCAGUGGCGGAGAAA-AAAGCCACCAGC ..-.(((((......)))))(((.(((((((((((.....))))))))))))))...........(((((...((((.......-...))))))))) ( -31.90) >consensus UUU__GUGUGUGUGUGUGCCGUGGCAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCCGGCAGUGGCGGAGAAA_AAAGCCACCAGC ..........((.(((.((.(((.(((((((((((.....))))))))))))))....((((((.(((......))))))))).....))))))).. (-25.96 = -25.52 + -0.44)

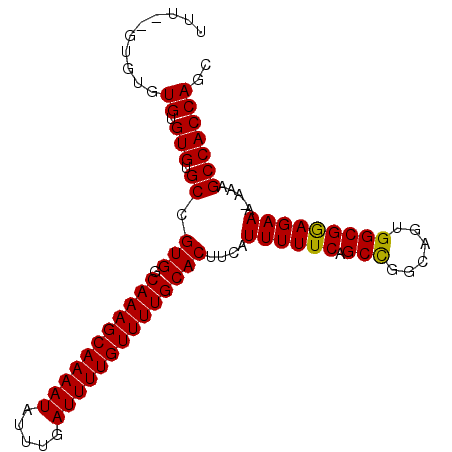

| Location | 11,344,425 – 11,344,519 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -22.21 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344425 94 - 27905053 GCUGGUGGCUUU-UUUCUUCGCCACUGCCAGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUGCCACGGCACACACACCCAC--AAA (((((((((...-.......)))...)))))).......((..(((((((.((((((.....)))))).)))).)))..))...........--... ( -22.40) >DroSec_CAF1 83293 95 - 1 GCUGGUGGCUUUUUUUCUCCGCCACUGCCGGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUGCCACGGCACACACACACAC--AAA ((((.(((((((.(((.((.(((......))).)).))).)))))(((((.((((((.....)))))).)))))))))))............--... ( -24.00) >DroSim_CAF1 81840 95 - 1 GCUGGUGGCUUUUUUUCUCCGCCACUGCCGGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUGCCACGGCACACACACACAC--AAA ((((.(((((((.(((.((.(((......))).)).))).)))))(((((.((((((.....)))))).)))))))))))............--... ( -24.00) >DroEre_CAF1 79547 95 - 1 GCUGGUGGCUUU-UUUCUCCGCCACUGCCAGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUGCCACGGCGCACACACACACCC-AA ((.((((((...-.......))))))(((.(((.........)))(((((.((((((.....)))))).)))))...)))))............-.. ( -23.60) >consensus GCUGGUGGCUUU_UUUCUCCGCCACUGCCAGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUGCCACGGCACACACACACAC__AAA (((((((((...........)))...)))))).......((..(((((((.((((((.....)))))).)))).)))..))................ (-22.21 = -21.77 + -0.44)



| Location | 11,344,447 – 11,344,559 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.84 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -33.84 |
| Energy contribution | -34.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344447 112 + 27905053 CAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCUGGCAGUGGCGAAGAAA-AAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAA (((((((((((.....))))))))))).(((((((.((((.((((((.(((((.......-...)))))-------..)))))).)...))).)))))))(((((.(....).))))).. ( -39.50) >DroSec_CAF1 83315 113 + 1 CAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCCGGCAGUGGCGGAGAAAAAAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAA (((((((((((.....))))))))))).(((((((.((((.((.(((.(((((...........)))))-------..))).)).)...))).)))))))(((((.(....).))))).. ( -34.80) >DroSim_CAF1 81862 113 + 1 CAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCCGGCAGUGGCGGAGAAAAAAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAA (((((((((((.....))))))))))).(((((((.((((.((.(((.(((((...........)))))-------..))).)).)...))).)))))))(((((.(....).))))).. ( -34.80) >DroEre_CAF1 79570 112 + 1 CAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCUGGCAGUGGCGGAGAAA-AAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAA (((((((((((.....))))))))))).(((((((.((((.((((((.(((((.......-...)))))-------..)))))).)...))).)))))))(((((.(....).))))).. ( -39.50) >DroYak_CAF1 81667 119 + 1 CAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUUAGCUGGCAGUGGCGGAGAAA-AAAGCCACCAGUUACCAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAA (((((((((((.....))))))))))).(((((((.((((.((((((.(((((.......-...)))))..(....).))))))....)))).)))))))(((((.(....).))))).. ( -37.90) >consensus CAAAGCAAAAUAUUUGAUUUUGUUUUGCACUUCAUUUUUCAGCUGGCAGUGGCGGAGAAA_AAAGCCAC_______CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAA (((((((((((.....))))))))))).(((((((.((((.((((((.(((((...........))))).........)))))).)...))).)))))))(((((.(....).))))).. (-33.84 = -34.08 + 0.24)



| Location | 11,344,447 – 11,344,559 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.84 |
| Mean single sequence MFE | -33.86 |
| Consensus MFE | -29.22 |
| Energy contribution | -29.18 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344447 112 - 27905053 UUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUU-UUUCUUCGCCACUGCCAGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUG (((((..((((..((..(((.......)))..))((((((((.(-------(((((...-.......))))))))))))))...))))..).))))..((((((.....))))))..... ( -34.70) >DroSec_CAF1 83315 113 - 1 UUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUUUUUUCUCCGCCACUGCCGGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUG (((((..((((..((..(((.......)))..))((((((((.(-------(((((...........))))))))))))))...))))..).))))..((((((.....))))))..... ( -33.90) >DroSim_CAF1 81862 113 - 1 UUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUUUUUUCUCCGCCACUGCCGGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUG (((((..((((..((..(((.......)))..))((((((((.(-------(((((...........))))))))))))))...))))..).))))..((((((.....))))))..... ( -33.90) >DroEre_CAF1 79570 112 - 1 UUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUU-UUUCUCCGCCACUGCCAGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUG (((((..((((..((..(((.......)))..))((((((((.(-------(((((...-.......))))))))))))))...))))..).))))..((((((.....))))))..... ( -34.70) >DroYak_CAF1 81667 119 - 1 UUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUGGUAACUGGUGGCUUU-UUUCUCCGCCACUGCCAGCUAAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUG ..(((...((((....)))).......((....)))))(((((((((..((((((....-.....)))))).)))))))))............((((.((((((.....)))))).)))) ( -32.10) >consensus UUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG_______GUGGCUUU_UUUCUCCGCCACUGCCAGCUGAAAAAUGAAGUGCAAAACAAAAUCAAAUAUUUUGCUUUG (((((..((((..((..(((.......)))..))((((((((.........(((((...........))))).))))))))...))))..).))))..((((((.....))))))..... (-29.22 = -29.18 + -0.04)


| Location | 11,344,487 – 11,344,599 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -47.54 |
| Consensus MFE | -43.56 |
| Energy contribution | -44.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344487 112 + 27905053 AGCUGGCAGUGGCGAAGAAA-AAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAAUCUGAUGGGCACGCGAGAGGGUUGCCCCGUCACCUCCUGA .((((((.(((((.......-...)))))-------..))))))...(((((.(((((.((((((.(....).)))))).)))(((((((((.(....).).)).)))))))).))))). ( -50.90) >DroSec_CAF1 83355 113 + 1 AGCCGGCAGUGGCGGAGAAAAAAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAAUCUGAUGGGCACGCGAGAGGGUUGCCCCGUCACCUCCUGA .((.(((.(((((...........)))))-------..))).))...(((((.(((((.((((((.(....).)))))).)))(((((((((.(....).).)).)))))))).))))). ( -46.20) >DroSim_CAF1 81902 113 + 1 AGCCGGCAGUGGCGGAGAAAAAAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAAUCUGAUGGGCACGCGAGAGGGUUGCCCCGUCACCUCCUGA .((.(((.(((((...........)))))-------..))).))...(((((.(((((.((((((.(....).)))))).)))(((((((((.(....).).)).)))))))).))))). ( -46.20) >DroEre_CAF1 79610 111 + 1 AGCUGGCAGUGGCGGAGAAA-AAAGCCAC-------CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAAUCUGAUGGGCACGCGAGCGGGUUGCCCCGUCACCUCC-GC .((((((.(((((.......-...)))))-------..)))))).........((((((((((((.(....).))))))...((((((((((.(....).).)).))))))).))))-)) ( -46.50) >DroYak_CAF1 81707 119 + 1 AGCUGGCAGUGGCGGAGAAA-AAAGCCACCAGUUACCAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAAUCUGAUGGGCACGCGAGAGGGUCGCUCCGUCACCUCCUGC .((((((.(((((.......-...)))))..(....).))))))...(((((.(((((.((((((.(....).)))))).)))(((((((((.(....).)).)).))))))).))))). ( -47.90) >consensus AGCUGGCAGUGGCGGAGAAA_AAAGCCAC_______CAGCCAGCCGCCAGGACGUGGAGUGUCAAGGAUAACGUUGGCAAUCUGAUGGGCACGCGAGAGGGUUGCCCCGUCACCUCCUGA .((((((.(((((...........))))).........))))))...(((((.(((((.((((((.(....).)))))).)))(((((((((.(....).)).)).))))))).))))). (-43.56 = -44.16 + 0.60)

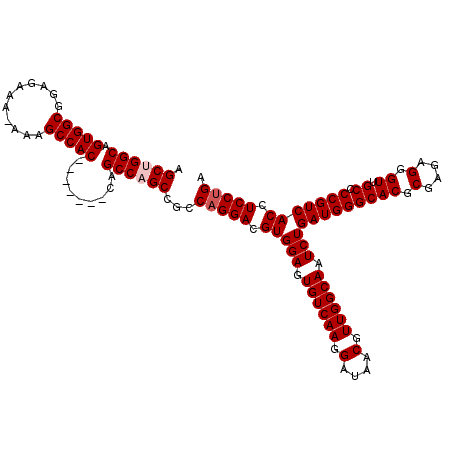

| Location | 11,344,487 – 11,344,599 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.29 |
| Mean single sequence MFE | -45.56 |
| Consensus MFE | -38.72 |
| Energy contribution | -38.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344487 112 - 27905053 UCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGAUUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUU-UUUCUUCGCCACUGCCAGCU ((((((.(((..(((....)))......(((..((...(((.((....)).)))..)).)))..))).)))))).(((((((.(-------(((((...-.......))))))))))))) ( -46.20) >DroSec_CAF1 83355 113 - 1 UCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGAUUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUUUUUUCUCCGCCACUGCCGGCU ((((((.(((..(((....)))......(((..((...(((.((....)).)))..)).)))..))).)))))).(((((((.(-------(((((...........))))))))))))) ( -45.40) >DroSim_CAF1 81902 113 - 1 UCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGAUUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUUUUUUCUCCGCCACUGCCGGCU ((((((.(((..(((....)))......(((..((...(((.((....)).)))..)).)))..))).)))))).(((((((.(-------(((((...........))))))))))))) ( -45.40) >DroEre_CAF1 79610 111 - 1 GC-GGAGGUGACGGGGCAACCCGCUCGCGUGCCCAUCAGAUUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG-------GUGGCUUU-UUUCUCCGCCACUGCCAGCU ..-((((((((((((....)))).))))....................((((....)))).)))).(((....)))((((((.(-------(((((...-.......)))))))))))). ( -47.60) >DroYak_CAF1 81707 119 - 1 GCAGGAGGUGACGGAGCGACCCUCUCGCGUGCCCAUCAGAUUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUGGUAACUGGUGGCUUU-UUUCUCCGCCACUGCCAGCU ((.((.((((.((((((((.....))))..(((.(((((.((((((..((((....))))...((((((....)))..))).))))))))))))))...-....)))).)))).)).)). ( -43.20) >consensus UCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGAUUGCCAACGUUAUCCUUGACACUCCACGUCCUGGCGGCUGGCUG_______GUGGCUUU_UUUCUCCGCCACUGCCAGCU .(((((.(((..(((....)))......(((..((...(((.((....)).)))..)).)))..))).)))))..(((((((.........(((((...........))))).))))))) (-38.72 = -38.88 + 0.16)



| Location | 11,344,559 – 11,344,661 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.56 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -27.72 |
| Energy contribution | -28.96 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344559 102 - 27905053 CUG------------------UUGCGGAUGCGACAAAAAUAAUGUAACGUCGGCGUGAACCUGUCACAAAGGAUGAUGGUUCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGA (((------------------.((.((((((((...............((((.(.((((((.((((.......))))))))))...).))))(((....)))..)))))).)))).))). ( -37.00) >DroSec_CAF1 83428 102 - 1 CUG------------------AUGCGGAUGCGACAAAAAUAAUGUAACGCCGGCGUGAACCUGUCACAAAGGAUGAUGGUUCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGA (((------------------(((.((((((((..............((((..(.((((((.((((.......)))))))))).).))))..(((....)))..)))))).)))))))). ( -44.20) >DroSim_CAF1 81975 102 - 1 CUG------------------AUGCGGAUGCGACAAAAAUAAUGUAACGCCGGCGUGAACCUGUCACAAAGGAUGAUGGCUCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGA (((------------------(((.((((((((..............((((..(.(((.((.((((.......)))))).))).).))))..(((....)))..)))))).)))))))). ( -40.00) >DroEre_CAF1 79682 119 - 1 CUGAUGCUGAUGCUGAUGCUGAUGCGGAUGCGACAAAAAUAAUGUAACGCCAGCGUGAACCUGUCACAAAGGAUGAUUGUGC-GGAGGUGACGGGGCAACCCGCUCGCGUGCCCAUCAGA ((((((.((.(((...(((....)))...))).))...........((((.(((((..((((.((((((.......))))).-).)))).))(((....)))))).))))...)))))). ( -41.00) >DroYak_CAF1 81786 120 - 1 CUGAUGCGGAUGCUGAUGCGGAUGCGGAUGCGACAAAAAUAAUGUAACGCCAGCGUGAACCUGUCACAAAGGAUGAUGGUGCAGGAGGUGACGGAGCGACCCUCUCGCGUGCCCAUCAGA ((((((.((((((((..(((..(((....)))(((.......)))..)))))))))....(((((((......((......))....))))))).((((.....))))...)))))))). ( -35.90) >consensus CUG__________________AUGCGGAUGCGACAAAAAUAAUGUAACGCCGGCGUGAACCUGUCACAAAGGAUGAUGGUUCAGGAGGUGACGGGGCAACCCUCUCGCGUGCCCAUCAGA .....................(((.((((((((..............((((..(.((((((.((((.......)))))))))).).))))..(((....)))..)))))).))))).... (-27.72 = -28.96 + 1.24)

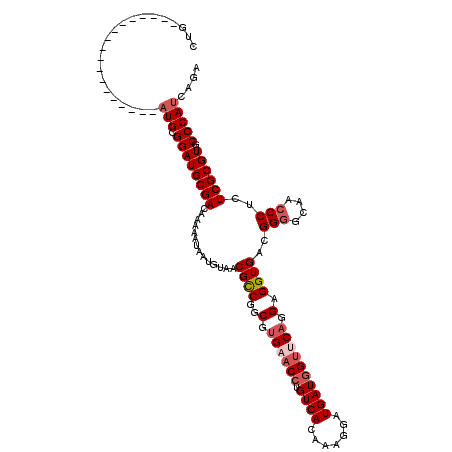

| Location | 11,344,661 – 11,344,769 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.98 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -23.80 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344661 108 + 27905053 CAUUCUCUUG----CCAACAAGGAAGCGGCAGCAGCAACAGGCCACAUCCCGCUCGAUGGGCU--------CAUCCCGGCUAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAA ..(((.((((----....)))))))..(((((((((....((((.((((......))))))))--------.......)))..)))))).............((((((.....)))))). ( -28.70) >DroSec_CAF1 83530 108 + 1 CAUUCGCUUG----CCAACAAGGAAGCUGGAGCAGCAACAGGCCACAUCCCGCUCGAUGGGCU--------CAUCCCGGCCAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAA ..(((.((((----....)))))))(((((.(((((....((((.((((......))))((..--------....))))))..)))))........))))).((((((.....)))))). ( -32.20) >DroSim_CAF1 82077 108 + 1 CAUUCGCUUG----CCAACAAGGAAGCGGGAGCAGCAACAGGCCACAUCCCGCUCGAUGGGCU--------CAUCCCGGCCAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAA .....((((.----((.....))))))((..(((((....((((.((((......))))((..--------....))))))..)))))..))..........((((((.....)))))). ( -33.30) >DroEre_CAF1 79801 108 + 1 CAUUCUCUUG----CCAACAAGGAACACGCAGCAGCAACAGGCCACGUCCUGCUCGAUGGGCU--------CAUCCCGGCUAGGCUGUCACCUUAACCGGCUUAAUGAACAACUCAUUAA .........(----((...((((.....((((((((....((((.((((......))))))))--------.......)))..)))))..))))....))).((((((.....)))))). ( -27.20) >DroYak_CAF1 81906 120 + 1 UAUUCUCUUCACCAUCAACAAGGAACAGGCAGCAGCAACAGGCCACAUCCUGCUCGAUGGGCUCAUUGACUCAUCCCGGCCAGGCUGUCACCUUAACCGGCUUAAUGAACAACUCAUUAA .....................((...(((..(((((....((((.(.....)...(((((((.....).))))))..))))..)))))..)))...))....((((((.....)))))). ( -28.70) >consensus CAUUCUCUUG____CCAACAAGGAAGCGGCAGCAGCAACAGGCCACAUCCCGCUCGAUGGGCU________CAUCCCGGCCAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAA ..(((.((((........)))))))..((..(((((....((((.((((......))))((..............))))))..)))))..))..........((((((.....)))))). (-23.80 = -24.00 + 0.20)

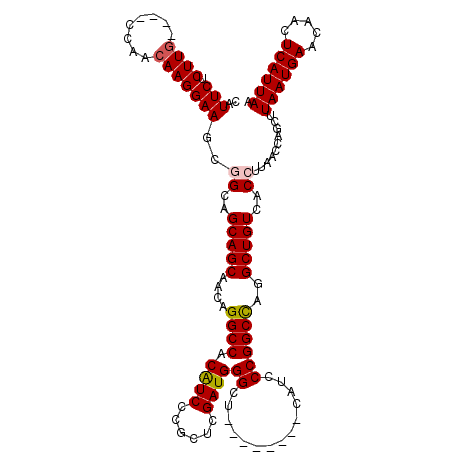
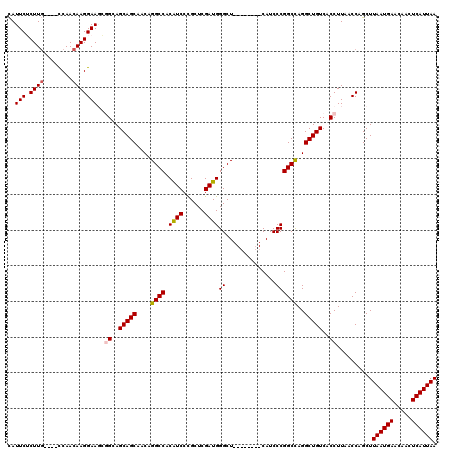
| Location | 11,344,697 – 11,344,809 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.74 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.48 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344697 112 + 27905053 GGCCACAUCCCGCUCGAUGGGCU--------CAUCCCGGCUAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAAUCAUGCACAUUUCGCAUUCAUAUACUUGCCGCGCGACAAA ((((.((((......))))))))--------..(((((((.((((((..........))))(((((((.....)))))))..((((.......)))).......)).)))).).)).... ( -26.80) >DroSec_CAF1 83566 112 + 1 GGCCACAUCCCGCUCGAUGGGCU--------CAUCCCGGCCAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAAUCAUGCACAUUUCGCAUUCAUAUACUUGCCGCGCGACAAA ((((.((((......))))))))--------..(((((((.((((((..........))))(((((((.....)))))))..((((.......)))).......)).)))).).)).... ( -26.80) >DroSim_CAF1 82113 112 + 1 GGCCACAUCCCGCUCGAUGGGCU--------CAUCCCGGCCAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAAUCAUGCACAUUUCGCAUUCAUAUACUUGCCUCGCGACAAA ((((.((((......))))))))--------.......((.((((..............(.(((((((.....))))))).)((((.......))))..........)))).))...... ( -24.90) >DroEre_CAF1 79837 110 + 1 GGCCACGUCCUGCUCGAUGGGCU--------CAUCCCGGCUAGGCUGUCACCUUAACCGGCUUAAUGAACAACUCAUUAAUCAUGCACAUUUCGCAUUCAUAUACUUGCC--GCGACAAA ((((.((((......))))))))--------..((.((((.((((((..........))))(((((((.....)))))))..((((.......)))).......)).)))--).)).... ( -26.20) >DroYak_CAF1 81946 118 + 1 GGCCACAUCCUGCUCGAUGGGCUCAUUGACUCAUCCCGGCCAGGCUGUCACCUUAACCGGCUUAAUGAACAACUCAUUAAUCAUGCACAUUUCGCAUUCAUAUACUAGCC--GCGACAAA ((..(((.((((((.(((((((.....).))))))..)).)))).)))..))....((((((((((((.....)))))))..((((.......))))..........)))--).)..... ( -29.80) >consensus GGCCACAUCCCGCUCGAUGGGCU________CAUCCCGGCCAGGCUGUCACCUUAACCAGCUUAAUGAACAACUCAUUAAUCAUGCACAUUUCGCAUUCAUAUACUUGCC_CGCGACAAA ((((.((((......))))))))..............(((.((((((..........))))(((((((.....)))))))..((((.......)))).......)).))).......... (-23.88 = -23.48 + -0.40)

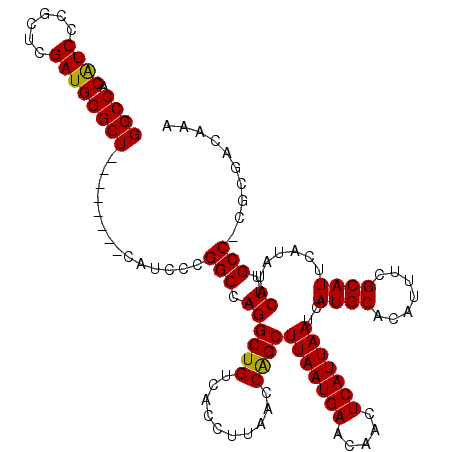

| Location | 11,344,697 – 11,344,809 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.74 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -32.08 |
| Energy contribution | -32.00 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11344697 112 - 27905053 UUUGUCGCGCGGCAAGUAUAUGAAUGCGAAAUGUGCAUGAUUAAUGAGUUGUUCAUUAAGCUGGUUAAGGUGACAGCCUAGCCGGGAUG--------AGCCCAUCGAGCGGGAUGUGGCC .(..(((((((....((((....))))....)))))....((((((((...)))))))).(((((((.(((....))))))))))))..--------)(((((((......)))).))). ( -36.20) >DroSec_CAF1 83566 112 - 1 UUUGUCGCGCGGCAAGUAUAUGAAUGCGAAAUGUGCAUGAUUAAUGAGUUGUUCAUUAAGCUGGUUAAGGUGACAGCCUGGCCGGGAUG--------AGCCCAUCGAGCGGGAUGUGGCC .(..(((((((....((((....))))....)))))....((((((((...)))))))).(((((((.(((....))))))))))))..--------)(((((((......)))).))). ( -35.80) >DroSim_CAF1 82113 112 - 1 UUUGUCGCGAGGCAAGUAUAUGAAUGCGAAAUGUGCAUGAUUAAUGAGUUGUUCAUUAAGCUGGUUAAGGUGACAGCCUGGCCGGGAUG--------AGCCCAUCGAGCGGGAUGUGGCC .(((((....)))))((((((.........))))))....((((((((...)))))))).(((((((.(((....))))))))))....--------.(((((((......)))).))). ( -34.80) >DroEre_CAF1 79837 110 - 1 UUUGUCGC--GGCAAGUAUAUGAAUGCGAAAUGUGCAUGAUUAAUGAGUUGUUCAUUAAGCCGGUUAAGGUGACAGCCUAGCCGGGAUG--------AGCCCAUCGAGCAGGACGUGGCC ...(((((--(.((.((((((.........)))))).)).........((((((......(((((((.(((....))))))))))((((--------....))))))))))..)))))). ( -36.10) >DroYak_CAF1 81946 118 - 1 UUUGUCGC--GGCUAGUAUAUGAAUGCGAAAUGUGCAUGAUUAAUGAGUUGUUCAUUAAGCCGGUUAAGGUGACAGCCUGGCCGGGAUGAGUCAAUGAGCCCAUCGAGCAGGAUGUGGCC ((..((.(--((((((.......(((((.....))))).........((((((......(((......)))))))))))))))).))..)).......(((((((......)))).))). ( -37.00) >consensus UUUGUCGCG_GGCAAGUAUAUGAAUGCGAAAUGUGCAUGAUUAAUGAGUUGUUCAUUAAGCUGGUUAAGGUGACAGCCUGGCCGGGAUG________AGCCCAUCGAGCGGGAUGUGGCC .(((((....)))))((((((.........))))))....((((((((...)))))))).(((((((.(((....)))))))))).............(((((((......)))).))). (-32.08 = -32.00 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:21:04 2006