
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,343,224 – 11,343,454 |
| Length | 230 |
| Max. P | 0.995654 |

| Location | 11,343,224 – 11,343,344 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -28.71 |
| Consensus MFE | -26.04 |
| Energy contribution | -25.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.91 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11343224 120 - 27905053 UGCGGAUUAACUUUUUACCAAUUUACAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAUAUUGAAAGUGUAAUAAAAAGCAGCUGUGCGCCGCUCACAC .((((....((.................((....))....((((((..(((......))).........((((((.(..........).))))))....))))))))...))))...... ( -26.40) >DroSec_CAF1 82075 120 - 1 UGCGGAUUAACUUUUAACCAAUUUAGAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAUAUUGAAAGUGUAAUAAAAAGCAGCUGUGCGCCGCUCACAC ...((.(((.....)))))......((((((...((....((((((..(((......))).........((((((.(..........).))))))....)))))).))..)))))).... ( -28.20) >DroSim_CAF1 80633 120 - 1 UGCGGAUUAACUUUUAACCAAUUUACAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAUAUUGAAAGUGUAAUAAAAAGCAGCUGUGCGCCGCUCACAC .((((....((.................((....))....((((((..(((......))).........((((((.(..........).))))))....))))))))...))))...... ( -26.40) >DroEre_CAF1 78371 120 - 1 UGCGGAUUAACUUUUAACCAAUUUACAGUGGAAGCAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUGCCCCACACUGAAAGUGUAAUAAAAAGCAGCUGUGCGCCGCUCCCAC .((((............(((........)))..(((....((((((..(((((.((((......)))).))..)))..((((.....))))........)))))).))).))))...... ( -32.20) >DroYak_CAF1 80403 120 - 1 UGCGGAUUAACUUUUAACCAAUUUACAGUGGAAGCAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACUCCAUACUGAAAGUGUAAUAAAAAGCAGCUGUGCGCCGCUCACAC .((((............(((........)))..(((....((((((..(((......))).........((((((..............))))))....)))))).))).))))...... ( -30.34) >consensus UGCGGAUUAACUUUUAACCAAUUUACAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAUAUUGAAAGUGUAAUAAAAAGCAGCUGUGCGCCGCUCACAC .((((....((......(((........))).........((((((..(((......))).........((((((..............))))))....))))))))...))))...... (-26.04 = -25.88 + -0.16)

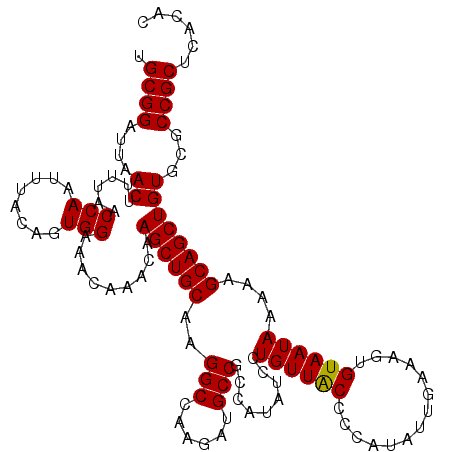

| Location | 11,343,264 – 11,343,384 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -31.34 |
| Energy contribution | -31.54 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11343264 120 - 27905053 GGCCUAUACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUUACCAAUUUACAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAU (((....((((((.....)))))).(((((.(......(((((((......(((((((.........)))))))........)))))))....).))))))))................. ( -30.64) >DroSec_CAF1 82115 120 - 1 GGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUAGAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAU (((...(((((((.....)))))))(((((.(......(((((((....(((((..........)))))(....).......)))))))....).))))))))................. ( -33.90) >DroSim_CAF1 80673 120 - 1 GGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAU (((...(((((((.....)))))))(((((.(......(((((((............(((........)))...........)))))))....).))))))))................. ( -32.00) >DroEre_CAF1 78411 120 - 1 GGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAGCAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUGCCCCAC (((...(((((((.....)))))))(((...........((((..............(((........))).(((......)))))))(((......))))))..........))).... ( -32.40) >DroYak_CAF1 80443 120 - 1 GGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAGCAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACUCCAU (((...(((((((.....)))))))(((((.(......(((((..............(((........))).(((......))))))))....).))))))))................. ( -32.10) >consensus GGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAACAAACAAGCUGCAAGGCCAAGAUGCCGCCAUAUCCUGUUACCCCAU (((...(((((((.....)))))))(((((.(......(((((((............(((........)))...........)))))))....).))))))))................. (-31.34 = -31.54 + 0.20)



| Location | 11,343,304 – 11,343,414 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.65 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11343304 110 - 27905053 ACGCGUGUC----AGCUCACGCACACUACUG------CACGGCCUAUACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUUACCAAUUUACAGUGGAAACAAACA ..(((((..----....)))))...((((((------.((((((...((((((.....)))))).))).)))..((((((.(..((.......))..).))))))))))))......... ( -33.10) >DroSec_CAF1 82155 114 - 1 ACGCGUGUCAGUCAGCUCUCGCACACUACUG------CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUAGAGUGGAAACAAACA .((((((.((((..((....)).....))))------))).(((..(((((((.....)))))))))).............))).....................(..((....))..). ( -30.40) >DroSim_CAF1 80713 114 - 1 ACGCGUGUCAGUCAGCUCACGCACACUACUG------CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAACAAACA ..(((((..((....)))))))...((((((------.((((((..(((((((.....)))))))))).)))...........((.(((.....)))))......))))))......... ( -31.30) >DroEre_CAF1 78451 97 - 1 -----------------CACGCACACUACUG------CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAGCAAACA -----------------...((...((((((------.((((((..(((((((.....)))))))))).)))...........((.(((.....)))))......))))))..))..... ( -25.30) >DroYak_CAF1 80483 103 - 1 -----------------CACGCACACUACUACGCACACACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAGCAAACA -----------------...((...(((((..(((...((((((..(((((((.....)))))))))).)))........)))((.(((.....))))).......)))))..))..... ( -23.90) >consensus ACGCGUGUC____AGCUCACGCACACUACUG______CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUUUGCGGAUUAACUUUUAACCAAUUUACAGUGGAAACAAACA .........................((((((.......((((((..(((((((.....)))))))))).)))..((((((.(..((.......))..).))))))))))))......... (-21.10 = -21.70 + 0.60)



| Location | 11,343,344 – 11,343,454 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11343344 110 - 27905053 AUGGCGGGAAAAUGAAAAGAGGCGGGAAAGCCUAACGCACACGCGUGUC----AGCUCACGCACACUACUG------CACGGCCUAUACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUU ((((((((....(((....((((......)))).((((....)))).))----).)))..(((......))------)...(((...((((((.....)))))).)))..)))))..... ( -36.90) >DroSec_CAF1 82195 114 - 1 AUGGCGGGGAAAUGAGAAGAGGCGGGAAAGCCUAAUACACACGCGUGUCAGUCAGCUCUCGCACACUACUG------CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUU ((((((((((..(((....((((......))))...((((....))))...)))..))))(((......))------)...(((..(((((((.....)))))))))).))))))..... ( -37.50) >DroSim_CAF1 80753 114 - 1 AUGGCGGGGAAAUGAAAAGAGGCGGGAAAGCCUAAUACACACGCGUGUCAGUCAGCUCACGCACACUACUG------CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUU ((((((.((..........((((......)))).........(((((..((....)))))))......)).------)...(((..(((((((.....))))))))))..)))))..... ( -36.00) >DroEre_CAF1 78491 89 - 1 GUGGCAGGAAAAUCAAAAG-GGCGAGAGAGCCU------------------------CACGCACACUACUG------CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUU ((((((............(-(((......))))------------------------...(((......))------)...(((..(((((((.....)))))))))).))))))..... ( -28.90) >DroYak_CAF1 80523 96 - 1 AUGGCAGUGAAAUCAAACGAGGCCAGAAUGCCU------------------------CACGCACACUACUACGCACACACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUU (((((((((.........(((((......))))------------------------)..((..........)).)))...(((..(((((((.....)))))))))).))))))..... ( -33.40) >consensus AUGGCGGGGAAAUGAAAAGAGGCGGGAAAGCCUAA__CACACGCGUGUC____AGCUCACGCACACUACUG______CACGGCCUAGACAGCCAAAAUGGCUGUUGGCAUGUCAUAAAUU ((((((.............((((......))))................................................(((..(((((((.....)))))))))).))))))..... (-25.72 = -25.72 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:54 2006