
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,340,599 – 11,341,039 |
| Length | 440 |
| Max. P | 0.999003 |

| Location | 11,340,599 – 11,340,713 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -32.54 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340599 114 - 27905053 CUUUGGCAUGUGUCGGCGUGCACCGAUAC------AUACAUAUCUAUGGUUCUGGGUAAGUGGGCCGUAGGGCCCGUCAGCCAGGUAAAAACAGGAAACAACUUUUGACCAUUUUUCAAA ...(((((((((((((......)))))))------))((.((((((......)))))).))(((((....)))))....))))(((.((((..(....)...)))).))).......... ( -38.20) >DroSec_CAF1 79393 114 - 1 CUUUGGCAUAUGUCGGCGUGCACCGAUAC------AUACAUAUCUAUGGCUCUGGGUAAGUGGGCCGUAGGGCCCGUCAGCCAGGUAAAAACAGGAAACAACUUUUGACCAUUUUUCAAA ...((((...((((((......)))))).------..((.((((((......)))))).))(((((....)))))....))))(((.((((..(....)...)))).))).......... ( -32.60) >DroSim_CAF1 77990 118 - 1 CUUUGGCAUAUGUCGGCGUGCACCGAUACACA--CAUACAUAUCUAUGGCUCUGGGUAAGUGGGCCGUAGGGCCCGUCAGCCAGGUAAAAACAGGAAACAACUUUUGACCAUUUUUCAAA ...((((...((((((......))))))....--...((.((((((......)))))).))(((((....)))))....))))(((.((((..(....)...)))).))).......... ( -32.60) >DroEre_CAF1 75110 110 - 1 CUUUGGCAUAUGUCGGCGUGCACCGAUAUG----------UAUCUAUGGCUCUGGGUAAGUGGGCCGUAGGGCCCGUCAGCCAGGUAAAAACAGGAAACAACUUUUGACCAUUUUUCAAA ...(((((((((((((......))))))))----------).((((((((((.........))))))))))........))))(((.((((..(....)...)))).))).......... ( -36.20) >DroYak_CAF1 76958 120 - 1 CUUUGGCAUAUGUCGGCGUGCACCGAUAUAUAUCCAUAUAUAUCUAUGGCUCUGGGUAAGUGGGCCGUAGGGCCCGUCAGCCAGGUAAAAACAGGAAACAACUUUUGACCAUUUUUCAAA ...(((((((((((((......)))))))))...........((((((((((.........))))))))))........))))(((.((((..(....)...)))).))).......... ( -36.20) >consensus CUUUGGCAUAUGUCGGCGUGCACCGAUAC______AUACAUAUCUAUGGCUCUGGGUAAGUGGGCCGUAGGGCCCGUCAGCCAGGUAAAAACAGGAAACAACUUUUGACCAUUUUUCAAA ...((((.((((((((......))))))))............((((((((((.........))))))))))........))))(((.((((..(....)...)))).))).......... (-32.54 = -32.94 + 0.40)



| Location | 11,340,639 – 11,340,753 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.80 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -27.06 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340639 114 + 27905053 CUGACGGGCCCUACGGCCCACUUACCCAGAACCAUAGAUAUGUAU------GUAUCGGUGCACGCCGACACAUGCCAAAGAUCACGCGCUGGCCCGAACAUAACCCAGUUUCAGAUCUCC .....(((((....)))))......................((((------((.(((((....))))).))))))...(((((....(((((............)))))....))))).. ( -33.70) >DroSec_CAF1 79433 112 + 1 CUGACGGGCCCUACGGCCCACUUACCCAGAGCCAUAGAUAUGUAU------GUAUCGGUGCACGCCGACAUAUGCCAAAGAUCACGCGCUGGCCCGAACAUAACCCAGUUCCAGAU--CC (((..(((((....)))))......((((.((....(((..((((------((.(((((....))))).)))))).....)))..)).))))...((((........)))))))..--.. ( -35.20) >DroSim_CAF1 78030 116 + 1 CUGACGGGCCCUACGGCCCACUUACCCAGAGCCAUAGAUAUGUAUG--UGUGUAUCGGUGCACGCCGACAUAUGCCAAAGAUCACGCGCUGGCCCGAACAUAACCCAGUUCCAGAU--CC (((..(((((....)))))......((((.((....(((..(((((--((....(((((....)))))))))))).....)))..)).))))...((((........)))))))..--.. ( -34.90) >DroEre_CAF1 75150 108 + 1 CUGACGGGCCCUACGGCCCACUUACCCAGAGCCAUAGAUA----------CAUAUCGGUGCACGCCGACAUAUGCCAAAGAUCACGCGCUGGCCCGAACAUAACCCAGUUCCAGAU--CC (((..(((((....)))))......((((.((....(((.----------(((((((((....)))))..))))......)))..)).))))...((((........)))))))..--.. ( -29.60) >DroYak_CAF1 76998 118 + 1 CUGACGGGCCCUACGGCCCACUUACCCAGAGCCAUAGAUAUAUAUGGAUAUAUAUCGGUGCACGCCGACAUAUGCCAAAGAUCACGCGCUGGCCCGAACAUAACCCAGUUCCAGAU--CC (((..(((((....)))))......((((.((....(((...(.(((.(((((.(((((....))))).)))))))).).)))..)).))))...((((........)))))))..--.. ( -36.50) >consensus CUGACGGGCCCUACGGCCCACUUACCCAGAGCCAUAGAUAUGUAU______GUAUCGGUGCACGCCGACAUAUGCCAAAGAUCACGCGCUGGCCCGAACAUAACCCAGUUCCAGAU__CC (((..(((((....)))))......((((.((....(((............((.(((((....))))).)).........)))..)).))))...((((........)))))))...... (-27.06 = -26.70 + -0.36)



| Location | 11,340,753 – 11,340,863 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -41.64 |
| Consensus MFE | -33.86 |
| Energy contribution | -33.70 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340753 110 + 27905053 GG----UUCUCCUUCGUG------GAGCACGUGAGUCCCGGCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCUCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCA .(----(.((((.....)------))).)).((..((((((((........(((((((..((..........))..)))).)))(((((......)))))))))))))..))........ ( -40.30) >DroSec_CAF1 79545 110 + 1 GG----UUCUCCUUCGUG------GAGCACGUGAGUCCCGGCUUAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCA .(----(.((((.....)------))).)).((..((((((((........(((...(((..(((........))).))).)))(((((......)))))))))))))..))........ ( -38.90) >DroSim_CAF1 78146 110 + 1 GG----UUCUCCUUCGUG------GAGCACGUGAGUCCCGGCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCA .(----(.((((.....)------))).)).((..((((((((........(((...(((..(((........))).))).)))(((((......)))))))))))))..))........ ( -39.50) >DroEre_CAF1 75258 116 + 1 GG----UUCUCCGUCGUGGGUACGGAGCACGUGAGUCCCGGCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCA .(----(.(((((((....).)))))).)).((..((((((((........(((...(((..(((........))).))).)))(((((......)))))))))))))..))........ ( -42.30) >DroYak_CAF1 77116 114 + 1 GGUCGGUGCUCCGUCGUG------GAGCACGUGAGUCCCGGCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGUCGGGAAGCAAGUUUCCA ((((((.(((((.....)------))))((....)).))))))....(((((.(((((.(((((((...((((...))))..))(((((......))))))))))))))))))))..... ( -47.20) >consensus GG____UUCUCCUUCGUG______GAGCACGUGAGUCCCGGCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCA ........................(((.((.((..((((((((........(((...(((..(((........))).))).)))(((((......)))))))))))))..)).))))).. (-33.86 = -33.70 + -0.16)

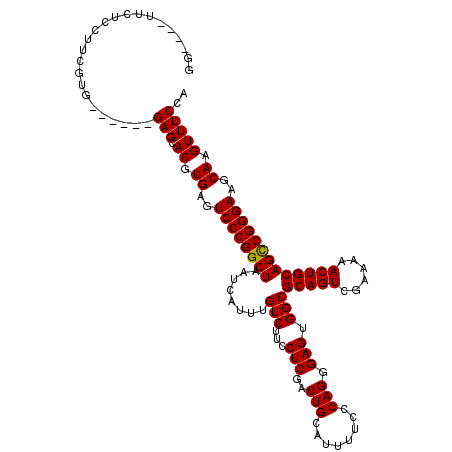

| Location | 11,340,753 – 11,340,863 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -41.03 |
| Consensus MFE | -37.16 |
| Energy contribution | -37.36 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340753 110 - 27905053 UGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGAGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGCCGGGACUCACGUGCUC------CACGAAGGAGAA----CC .(....).((..(((((((((...(((((((((((((...(((...)))....)))))))))))))..(....)..)))))))))..)).((.(((------(.....)))).)----). ( -39.80) >DroSec_CAF1 79545 110 - 1 UGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUAAGCCGGGACUCACGUGCUC------CACGAAGGAGAA----CC .(....).((..((((((((.((.(((((((((((((....(((...)))...)))))))))))))......))...))))))))..)).((.(((------(.....)))).)----). ( -41.30) >DroSim_CAF1 78146 110 - 1 UGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGCCGGGACUCACGUGCUC------CACGAAGGAGAA----CC .(....).((..(((((((((...(((((((((((((....(((...)))...)))))))))))))..(....)..)))))))))..)).((.(((------(.....)))).)----). ( -40.30) >DroEre_CAF1 75258 116 - 1 UGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGCCGGGACUCACGUGCUCCGUACCCACGACGGAGAA----CC .(....).((..(((((((((...(((((((((((((....(((...)))...)))))))))))))..(....)..)))))))))..)).((.((((((.......)))))).)----). ( -43.70) >DroYak_CAF1 77116 114 - 1 UGGAAACUUGCUUCCCGACUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGCCGGGACUCACGUGCUC------CACGACGGAGCACCGACC .(....)((((((((((((((((.(((..((..............))..))).)))))))).))))).)))...................((((((------(.....)))))))..... ( -40.04) >consensus UGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGCCGGGACUCACGUGCUC______CACGAAGGAGAA____CC .(....).....((((((((.((.(((((((((((((....((.....))...)))))))))))))......))...))))))))(((.((((.........))))...)))........ (-37.16 = -37.36 + 0.20)



| Location | 11,340,783 – 11,340,903 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.923401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340783 120 + 27905053 GCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCUCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCAUUACGCACUGACAGUUGAUUGCUAAGUUUAUUAUUGCUUU ((.(((((.(((((....((((((((..((((....))))....)))))))).....(((.....(((((....))))).....)))..))))).))))))).((((........)))). ( -33.40) >DroSec_CAF1 79575 120 + 1 GCUUAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCAUUACGCACUGACAGUUGAUUGCUAAGUUUAUUAUUGCUUU ((..((((.(((((....((((((((...((((...))))....)))))))).....(((.....(((((....))))).....)))..))))).)))).)).((((........)))). ( -33.50) >DroSim_CAF1 78176 120 + 1 GCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCAUUACGCACUGACAGUUGAUUGCUAAGUUUAUUAUUGCUUU ((.(((((.(((((....((((((((...((((...))))....)))))))).....(((.....(((((....))))).....)))..))))).))))))).((((........)))). ( -34.70) >DroEre_CAF1 75294 120 + 1 GCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCAUUACGCACUGACAGUUGAUUGCUAAGUUUAUUAUUGCUUU ((.(((((.(((((....((((((((...((((...))))....)))))))).....(((.....(((((....))))).....)))..))))).))))))).((((........)))). ( -34.70) >DroYak_CAF1 77150 120 + 1 GCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGUCGGGAAGCAAGUUUCCAUUACGCACUGACAGUUGAUUGCUAAGUUUAUUAUUGCUUU ((.(((((.(((((....((((((((...((((...))))....)))))))).....(((.((..(((((....)))))...)))))..))))).))))))).((((........)))). ( -35.00) >consensus GCUAAUCAUUUGUCUUCCUCGACUGCAUUUUCCCAGGGAGUGGCGCAGUCGAAAAACUGCAGCCGGGAAGCAAGUUUCCAUUACGCACUGACAGUUGAUUGCUAAGUUUAUUAUUGCUUU ((.(((((.(((((....((((((((...((((...))))....)))))))).....(((.....(((((....))))).....)))..))))).))))))).((((........)))). (-33.72 = -33.76 + 0.04)


| Location | 11,340,783 – 11,340,903 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -34.68 |
| Energy contribution | -34.56 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.999003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340783 120 - 27905053 AAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGAGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGC ..................(((((((..((((.....((((.(....)))))((((((((((((.(((((((..............))))))).)))))))).))))))))..))))).)) ( -35.84) >DroSec_CAF1 79575 120 - 1 AAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUAAGC ...((.............)).((((..((((.....((((.(....)))))((((((((((((.(((..((..............))..))).)))))))).))))))))..)))).... ( -32.76) >DroSim_CAF1 78176 120 - 1 AAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGC ..................(((((((..((((.....((((.(....)))))((((((((((((.(((..((..............))..))).)))))))).))))))))..))))).)) ( -35.04) >DroEre_CAF1 75294 120 - 1 AAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGC ..................(((((((..((((.....((((.(....)))))((((((((((((.(((..((..............))..))).)))))))).))))))))..))))).)) ( -35.04) >DroYak_CAF1 77150 120 - 1 AAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGACUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGC ..................(((((((..((((.....((((.(....)))))((((((((((((.(((..((..............))..))).)))))))).))))))))..))))).)) ( -35.64) >consensus AAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCACUCCCUGGGAAAAUGCAGUCGAGGAAGACAAAUGAUUAGC ..................(((((((..((((.....((((.(....)))))((((((((((((.(((((((..............))))))).)))))))).))))))))..))))).)) (-34.68 = -34.56 + -0.12)



| Location | 11,340,823 – 11,340,919 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340823 96 - 27905053 UA------------------------CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCA ..------------------------.(((((((((.((....)).............(((......)))...)))))...(....)....)))).(((.((((((....))))))))). ( -28.40) >DroSec_CAF1 79615 96 - 1 UA------------------------CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCA ..------------------------.(((((((((.((....)).............(((......)))...)))))...(....)....)))).(((.((((((....))))))))). ( -28.40) >DroSim_CAF1 78216 96 - 1 UA------------------------CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCA ..------------------------.(((((((((.((....)).............(((......)))...)))))...(....)....)))).(((.((((((....))))))))). ( -28.40) >DroEre_CAF1 75334 120 - 1 UACGGAUACGGAUGCGGAUGCGGAUGCGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCA ...((((((.((((..(((((...((((....)))).((....)).............)).)))..).))).))).((((.(....))))).))).(((.((((((....))))))))). ( -40.30) >DroYak_CAF1 77190 114 - 1 UA------CGGAUGCGGAUGCGGAUACGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGACUGCAGUUUUUCGACUGCGCCA ..------.((.(((((.(((.....((....))...((....)).............)))...(((((.((((.((....((((......)))))))))))))))......))))))). ( -33.00) >consensus UA________________________CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAAUGGAAACUUGCUUCCCGGCUGCAGUUUUUCGACUGCGCCA ...........................(((((((((.((....)).............(((......)))...)))))...(....)....)))).(((.((((((....))))))))). (-26.86 = -27.06 + 0.20)



| Location | 11,340,863 – 11,340,959 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -26.54 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340863 96 - 27905053 AUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGAUA------------------------CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAA .(((...(((.(((......))))))....((....))....------------------------.))).(((((.((....)).............(((......)))...))))).. ( -23.20) >DroSec_CAF1 79655 96 - 1 AUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGAUA------------------------CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAA .(((...(((.(((......))))))....((....))....------------------------.))).(((((.((....)).............(((......)))...))))).. ( -23.20) >DroSim_CAF1 78256 96 - 1 AUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGAUA------------------------CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAA .(((...(((.(((......))))))....((....))....------------------------.))).(((((.((....)).............(((......)))...))))).. ( -23.20) >DroEre_CAF1 75374 120 - 1 AUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGAUACGGAUACGGAUGCGGAUGCGGAUGCGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAA .((((((........)))).))(((((..((((...((.((.((....)).)).))(((((...((((....)))).((....)).............)).)))..))))...))))).. ( -34.60) >DroYak_CAF1 77230 114 - 1 AUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGAUA------CGGAUGCGGAUGCGGAUACGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAA .((((((........)))).))(((((...((....))((((------..((((((..(.((....)).)..)))..((....))................)))...))))..))))).. ( -28.50) >consensus AUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGAUA________________________CGGAAACGCAAGCGAAAGCAAUAAUAAACUUAGCAAUCAACUGUCAGUGCGUAA .(((...(((.(((......))))))....((....)).............................))).(((((.((....)).............(((......)))...))))).. (-23.20 = -23.20 + -0.00)



| Location | 11,340,919 – 11,341,039 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -31.50 |
| Consensus MFE | -30.16 |
| Energy contribution | -30.08 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11340919 120 - 27905053 AAAAGAUAAGUUGGAAAACGCUGCGAGCUUCCAUAAUGCCAUCAAAGGUCAACACUUGAUAAAUAGUUGCAGCUCGGCGAAUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGA ....(((..(((((.....(((((((((.........)).(((((..(....)..)))))......))))))))))))..)))(((.(((.(((......)))))).)))((....)).. ( -26.80) >DroSec_CAF1 79711 120 - 1 AAAGGAUAAGUUGGGAAACGCUGCGAGCUUCCAUAAUGCCAUCAAAGGUCAACACUUGAUAAAUAGUUGCAGCUCGGCGAAUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGA .((((((..(((((....)(((((((((.........)).(((((..(....)..)))))......))))))).))))..)))))).(((.(((......))))))....((....)).. ( -34.50) >DroSim_CAF1 78312 120 - 1 AAAGGAUAAGUUGGGAAACGCUGCGAGCUUCCAUAAUGCCAUCAAAGGUCAACACUUGAUAAAUAGUUGCAGCUCGGCGAAUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGA .((((((..(((((....)(((((((((.........)).(((((..(....)..)))))......))))))).))))..)))))).(((.(((......))))))....((....)).. ( -34.50) >DroEre_CAF1 75454 120 - 1 AAGGGAUAAGUUGGGAAAUGCUGCGAGCUUCCAUAAUGCCAUCAGAGGUCAACACUUGAUAAAUAGUUGCAGCUCGGCGAAUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGA .((((((..((((((...(((.(((.((.........)))(((((..(....)..))))).....)).))).))))))..)))))).(((.(((......))))))....((....)).. ( -31.60) >DroYak_CAF1 77304 120 - 1 AAAGGGUAAGUUGUAAAACGCUGCGAGCUUCCAUAAUGCCAUCAAAGGUCAACACUUGAUAAAUAGUUGCAGCUCGGCGAAUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGA .((((((..((((......(((((((((.........)).(((((..(....)..)))))......))))))).))))..)))))).(((.(((......))))))....((....)).. ( -30.10) >consensus AAAGGAUAAGUUGGGAAACGCUGCGAGCUUCCAUAAUGCCAUCAAAGGUCAACACUUGAUAAAUAGUUGCAGCUCGGCGAAUCCUUGGCGAUUCAUAAGCGAACGCAAAGCGGAAACGGA .((((((..(((((.....(((((((((.........)).(((((..(....)..)))))......))))))))))))..)))))).(((.(((......))))))....((....)).. (-30.16 = -30.08 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:45 2006