
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,338,205 – 11,338,456 |
| Length | 251 |
| Max. P | 0.903095 |

| Location | 11,338,205 – 11,338,313 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.74 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.66 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11338205 108 + 27905053 CGU------------UCUCAUCCUUUAUUGUUUUCCGUGUUCAGUAGCUGUCUUCUACUCCUGGUCCUCGAGGGUGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAUUCU ...------------.....................((((.(((((...(....(((....)))....)((((((((((((((...))))))))))))))........)))))))))... ( -28.40) >DroSec_CAF1 77028 108 + 1 CGU------------CCUCAUCCUUUAUUGUUUUCCGUGUUCAGUAGCUGUCUUCUACUCGUGGUUCUCGAGGGAGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCU ...------------..........(((((..(.....)..)))))((((((..(((....))).....(((((.((((((((...)))))))).)))))............)))))).. ( -25.90) >DroSim_CAF1 75617 108 + 1 CGU------------CCUCAUCCUUUAUUGUUUUCCGUGUUCAGUAGCUGUCUUCUACUCCUGGUCCUCGAGGGUGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCU ...------------..........(((((..(.....)..)))))((((((..(((....))).....((((((((((((((...))))))))))))))............)))))).. ( -29.30) >DroEre_CAF1 72913 120 + 1 CAUCCUCGUCCUCAUGCUCAUCCUUUAUUGUUUUCCUUUCUCAGUAGCUGUCUGUUUCUCCUGGUCCUCGAGGGUGCGAGUACGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCU .........................(((((...........)))))((((((.(..((....))..)..((((((((((((.......))))))))))))............)))))).. ( -24.50) >DroYak_CAF1 74640 108 + 1 CAC------------UCUCAUCCUUUAUUGUUUUCCAUUCUCAGUAGCUGUCUUUUUCUCCUGGUCUUCGAGGGUGCGAGUACGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCU ...------------..........(((((...........)))))((((((....((....)).....((((((((((((.......))))))))))))............)))))).. ( -23.40) >consensus CGU____________CCUCAUCCUUUAUUGUUUUCCGUGUUCAGUAGCUGUCUUCUACUCCUGGUCCUCGAGGGUGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCU .........................(((((...........)))))((((((.....(....)......((((((((((((((...))))))))))))))............)))))).. (-23.86 = -24.66 + 0.80)



| Location | 11,338,233 – 11,338,353 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -28.77 |
| Energy contribution | -29.57 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11338233 120 + 27905053 UCAGUAGCUGUCUUCUACUCCUGGUCCUCGAGGGUGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAUUCUGAGUCCGGAACUUCAACCCUCGUUCCCCUGACUUUUGCUC .(((((...(....(((....)))....)((((((((((((((...))))))))))))))........)))))(((....(((((.(((((..........)))))...))))).))).. ( -38.60) >DroSec_CAF1 77056 120 + 1 UCAGUAGCUGUCUUCUACUCGUGGUUCUCGAGGGAGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCUGAGUCCGGAACUUCAACCCUCGUCCCCCUGACUUUUGCUC ..(((((..(((.......((.((((...(((((.((((((((...)))))))).)))))..........(((((.......).))))......))))..)).......)))..))))). ( -32.74) >DroSim_CAF1 75645 120 + 1 UCAGUAGCUGUCUUCUACUCCUGGUCCUCGAGGGUGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCUGAGUCCGGAACUUCAACCCUCGUCCCCCCGACUUUUGCUC ((((..((((((..(((....))).....((((((((((((((...))))))))))))))............))))))))))(((.((..................)).)))........ ( -35.17) >DroEre_CAF1 72953 119 + 1 UCAGUAGCUGUCUGUUUCUCCUGGUCCUCGAGGGUGCGAGUACGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCUGCGUCCGGAACAUCAACCCUCGUUC-CCCGACUUUUGCUC ...(((((((((.(..((....))..)..((((((((((((.......))))))))))))............))))).))))(((.(((((..........))))-)..)))........ ( -35.80) >DroYak_CAF1 74668 120 + 1 UCAGUAGCUGUCUUUUUCUCCUGGUCUUCGAGGGUGCGAGUACGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCUGCGUCCUGAACUUCAACCCUCGUCCCCCCGACUUUUGCUC ...(((((((((....((....)).....((((((((((((.......))))))))))))............))))).))))...................(((.....)))........ ( -28.70) >consensus UCAGUAGCUGUCUUCUACUCCUGGUCCUCGAGGGUGCGAGUGCGUCGCAUUUGCACCUUCAAUUAAUAUACUGGCAGUCUGAGUCCGGAACUUCAACCCUCGUCCCCCCGACUUUUGCUC ..(((((..(((..........((.....((((((((((((((...))))))))))))))..........(((((.......).)))).........))..........)))..))))). (-28.77 = -29.57 + 0.80)

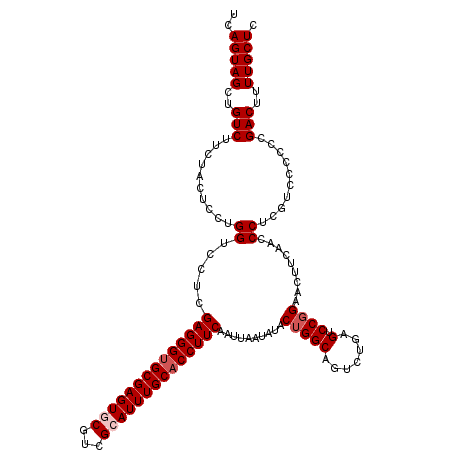

| Location | 11,338,353 – 11,338,456 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.26 |
| Mean single sequence MFE | -32.78 |
| Consensus MFE | -32.06 |
| Energy contribution | -31.58 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11338353 103 + 27905053 UU-------UUUGUGAUUUAUGAGGGUGCGCUGGCAAAACAAAAUUAAGCCGGCGGACGAACGACAUUUGGCGUCAGCAUCUUAGCCGGGCUUUCCCAGC------AACCACAUCC---- ..-------..((((.......(((((((((((((.............)))))).((((..(((...))).)))).))))))).((.(((....))).))------...))))...---- ( -32.72) >DroSec_CAF1 77176 103 + 1 UU-------UUUGUGAUUUAUGAGGGUGCGCUGGCAAAACAAAAUUAAGCCGGCGGACGAACGACAUUCGGCGUCAGCAUCUUAGCGGGGCUUUCCCAGU------AACCACAUCC---- ..-------..((((.......(((((((((((((.............)))))).((((..((.....)).)))).)))))))...(((.....)))...------...))))...---- ( -32.92) >DroSim_CAF1 75765 103 + 1 UU-------UUUGUGAUUUAUGAGGGUGCGCUGGCAAAACAAAAUUAAGCCGGCGGACGAACGACAUUCGGCGUCAGCAUCUUAGCCGGGCUUUCCCAGU------AACCACAUCC---- ..-------..((((.......(((((((((((((.............)))))).((((..((.....)).)))).))))))).((.(((....))).))------...))))...---- ( -32.82) >DroEre_CAF1 73072 103 + 1 CU-------UUUGUGAUUUAUGAGGGUGCGCUGGCAAAACAAAAUUAAGCCGGCGGACGAACGACACUUGGCGUCAGCAUCUUAGCCGGGCUUUCCCAGC------AACCACAUCC---- ..-------..((((.......(((((((((((((.............)))))).((((..(((...))).)))).))))))).((.(((....))).))------...))))...---- ( -32.72) >DroYak_CAF1 74788 120 + 1 UUUUCGCUUUUUGUGAUUUAUGAGGGUGCGCUGGCAAAACAAAAUUAAGCCGGCGGACGAACGACAUUUGGCGUCAGCAUCUUAGCCGGGCUCUCCCAGCUUUUCCAACCACAUCCCAUC ...........((((.......(((((((((((((.............)))))).((((..(((...))).)))).)))))))(((.(((....))).)))........))))....... ( -32.72) >consensus UU_______UUUGUGAUUUAUGAGGGUGCGCUGGCAAAACAAAAUUAAGCCGGCGGACGAACGACAUUUGGCGUCAGCAUCUUAGCCGGGCUUUCCCAGC______AACCACAUCC____ ...........((((.......(((((((((((((.............)))))).((((..(((...))).)))).))))))).((.(((....))).)).........))))....... (-32.06 = -31.58 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:35 2006