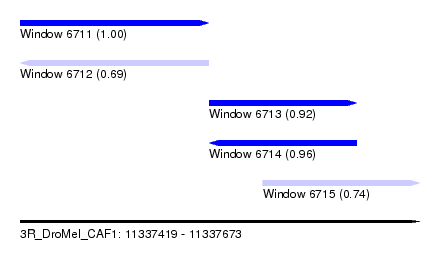
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,337,419 – 11,337,673 |
| Length | 254 |
| Max. P | 0.999789 |

| Location | 11,337,419 – 11,337,539 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -38.46 |
| Energy contribution | -38.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11337419 120 + 27905053 GCUUUUGUAGCUGCAAGGAUAUUGCAGCUGCAACGUGUUGAAGUGGCGCCUCGGUAAAAUUGUGUCAAACUGUGAUUGCAUUUUUGUGACUGGCAUUCAAUUGCUAUGGUGUUUGGCUUU ....((((((((((((.....))))))))))))......(((((((((((..(((((....((((((.((.(((....)))....))...))))))....)))))..))))))..))))) ( -38.90) >DroSec_CAF1 76243 120 + 1 GCUUUUGUAGCUGCAAGGAUAUUGCAGCUGCAACGUGUUGAAGUGGCGCCUCGGUAAAAUUGUGUCAAACUGUGAUUGCAUUUUUGUGACUGGCAUUCAAUUGCUGUGGUGUUUGGCUUU ....((((((((((((.....))))))))))))......(((((((((((.((((((....((((((.((.(((....)))....))...))))))....)))))).))))))..))))) ( -41.80) >DroSim_CAF1 74841 120 + 1 GCUUUUGUAGCUGCAAGGAUAUUGCAGCUGCAACGUGUUGAAGUGGCGCCUCGGUAAAAUUGUGUCAAACUGUGAUUGCAUUUUUGUGACUGGCAUUCAAUUGCUGUGGUGUUUGGCUUU ....((((((((((((.....))))))))))))......(((((((((((.((((((....((((((.((.(((....)))....))...))))))....)))))).))))))..))))) ( -41.80) >DroEre_CAF1 72132 120 + 1 GCUCUUGUAACUGCAAGGAUAUUGCAGCUGCAACGUGUUGAAGUGGCGCCUCGGUGAAAUUGUGUCAAACUGUGAUUGCAUUUUUGUGACUGGCAUUCAAUUGCUGUGGUGUUUGGCUUU ((.((((((.((((((.....)))))).))))).).)).(((((((((((.(((..(....((((((.((.(((....)))....))...))))))....)..))).))))))..))))) ( -35.20) >DroYak_CAF1 73854 120 + 1 GCUUUUGUAGCUGUAAGGAUAUUGCAGCUGCAACGUGUUGAAGUGGCGCCUCGGUAAAAUUGUGUCAAACUGUGAUUGCAUUUUUGUGACUGCCAUUCAAUUGCUGUGGUGUUUGGCUUU ....((((((((((((.....))))))))))))......(((((((((((.(((((((((.((((((....(((....))).....)))).)).)))...)))))).))))))..))))) ( -37.60) >consensus GCUUUUGUAGCUGCAAGGAUAUUGCAGCUGCAACGUGUUGAAGUGGCGCCUCGGUAAAAUUGUGUCAAACUGUGAUUGCAUUUUUGUGACUGGCAUUCAAUUGCUGUGGUGUUUGGCUUU ....((((((((((((.....))))))))))))......(((((((((((.((((((....((((((.((.(((....)))....))...))))))....)))))).))))))..))))) (-38.46 = -38.74 + 0.28)


| Location | 11,337,419 – 11,337,539 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -22.32 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.84 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.689869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11337419 120 - 27905053 AAAGCCAAACACCAUAGCAAUUGAAUGCCAGUCACAAAAAUGCAAUCACAGUUUGACACAAUUUUACCGAGGCGCCACUUCAACACGUUGCAGCUGCAAUAUCCUUGCAGCUACAAAAGC ...(((..........(((......)))..((((.((...((......)).)))))).............)))((.((........)).))((((((((.....))))))))........ ( -23.40) >DroSec_CAF1 76243 120 - 1 AAAGCCAAACACCACAGCAAUUGAAUGCCAGUCACAAAAAUGCAAUCACAGUUUGACACAAUUUUACCGAGGCGCCACUUCAACACGUUGCAGCUGCAAUAUCCUUGCAGCUACAAAAGC ...(((..........(((......)))..((((.((...((......)).)))))).............)))((.((........)).))((((((((.....))))))))........ ( -23.40) >DroSim_CAF1 74841 120 - 1 AAAGCCAAACACCACAGCAAUUGAAUGCCAGUCACAAAAAUGCAAUCACAGUUUGACACAAUUUUACCGAGGCGCCACUUCAACACGUUGCAGCUGCAAUAUCCUUGCAGCUACAAAAGC ...(((..........(((......)))..((((.((...((......)).)))))).............)))((.((........)).))((((((((.....))))))))........ ( -23.40) >DroEre_CAF1 72132 120 - 1 AAAGCCAAACACCACAGCAAUUGAAUGCCAGUCACAAAAAUGCAAUCACAGUUUGACACAAUUUCACCGAGGCGCCACUUCAACACGUUGCAGCUGCAAUAUCCUUGCAGUUACAAGAGC ...(((..........(((......)))..((((.((...((......)).)))))).............)))((.((........)).))((((((((.....))))))))........ ( -21.00) >DroYak_CAF1 73854 120 - 1 AAAGCCAAACACCACAGCAAUUGAAUGGCAGUCACAAAAAUGCAAUCACAGUUUGACACAAUUUUACCGAGGCGCCACUUCAACACGUUGCAGCUGCAAUAUCCUUACAGCUACAAAAGC ................(((((((((((((.(((.(.(((((....(((.....)))....)))))...).))))))).))))....)))))(((((...........)))))........ ( -20.40) >consensus AAAGCCAAACACCACAGCAAUUGAAUGCCAGUCACAAAAAUGCAAUCACAGUUUGACACAAUUUUACCGAGGCGCCACUUCAACACGUUGCAGCUGCAAUAUCCUUGCAGCUACAAAAGC ...(((..........(((......)))..((((.((...((......)).)))))).............)))((.((........)).))((((((((.....))))))))........ (-20.60 = -20.84 + 0.24)



| Location | 11,337,539 – 11,337,633 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -24.96 |
| Energy contribution | -24.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11337539 94 + 27905053 GGCAACUCUGCCCAUGUCCUUAACUCCCUUUACC----------------------CGCCCACUUGG----GCAUGUGGGCGUGGUCCGAUUUGGGCACUAACACUUGUCGCCUUCAGAU (((.....((((((.(((.............(((----------------------(((((((....----....))))))).)))..))).))))))...((....)).)))....... ( -31.46) >DroSec_CAF1 76363 94 + 1 GGCAACUCUGCCCAUGUCCUCAACUCCCUUUACC----------------------CGCCCACUUGG----GCAUGUGGGCGUGGUCCGAUUUAGGCACUAACACUUGUCGCCUUCAGAU ((((....))))...................(((----------------------(((((((....----....))))))).)))..((...((((((........)).)))))).... ( -28.30) >DroSim_CAF1 74961 94 + 1 GGCAACUCUGCCCAUGUCCUCAACUCCCUUUACC----------------------CGCCCACUUGG----GCAUGUGGGCGUGGUCCGAUUUGGGCCCUAACACUUGUCGCCUUCAGAU (((......(((((.(((.............(((----------------------(((((((....----....))))))).)))..))).)))))....((....)).)))....... ( -30.06) >DroEre_CAF1 72252 120 + 1 GGCAACCCUGCCCAAGUGCUCAACUCCCUUUUCCCAGAAUACCUACACUCCACUGCCACCCACUUGGGCGGCCAUGUGGGCGUGGUCCGAUUUGGGCACUAACACUUGUCGCCCUUGGAC ((((....)))).................................((((((((.(((.(((....))).)))...))))).)))((((((...((((((........)).)))))))))) ( -39.70) >DroYak_CAF1 73974 87 + 1 GGCAACCCUGCCCAUGUCCUCCACUCC-----------------------------CGCCCACUUGG----UCAUGUGGGCGUGGUCCGAAGUGGGCACUAACACUUGUCGCCUUCGGAC ((((....)))).............((-----------------------------(((((((....----....))))))).))(((((((..((((........))))..))))))). ( -35.00) >consensus GGCAACUCUGCCCAUGUCCUCAACUCCCUUUACC______________________CGCCCACUUGG____GCAUGUGGGCGUGGUCCGAUUUGGGCACUAACACUUGUCGCCUUCAGAU ((((....))))............................................(((((((.((......)).)))))))..(((.((...((((((........)).)))))).))) (-24.96 = -24.64 + -0.32)



| Location | 11,337,539 – 11,337,633 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -28.06 |
| Energy contribution | -27.94 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11337539 94 - 27905053 AUCUGAAGGCGACAAGUGUUAGUGCCCAAAUCGGACCACGCCCACAUGC----CCAAGUGGGCG----------------------GGUAAAGGGAGUUAAGGACAUGGGCAGAGUUGCC .......((((((.........((((((..((..(((.(((((((....----....)))))))----------------------)))...))..(((...))).))))))..)))))) ( -33.80) >DroSec_CAF1 76363 94 - 1 AUCUGAAGGCGACAAGUGUUAGUGCCUAAAUCGGACCACGCCCACAUGC----CCAAGUGGGCG----------------------GGUAAAGGGAGUUGAGGACAUGGGCAGAGUUGCC .......((((((...(((..((((((...((..(((.(((((((....----....)))))))----------------------)))...))......))).)))..)))..)))))) ( -36.40) >DroSim_CAF1 74961 94 - 1 AUCUGAAGGCGACAAGUGUUAGGGCCCAAAUCGGACCACGCCCACAUGC----CCAAGUGGGCG----------------------GGUAAAGGGAGUUGAGGACAUGGGCAGAGUUGCC .......((((((...((((...((((((.((..(((.(((((((....----....)))))))----------------------)))...))...))).)).)...))))..)))))) ( -35.20) >DroEre_CAF1 72252 120 - 1 GUCCAAGGGCGACAAGUGUUAGUGCCCAAAUCGGACCACGCCCACAUGGCCGCCCAAGUGGGUGGCAGUGGAGUGUAGGUAUUCUGGGAAAAGGGAGUUGAGCACUUGGGCAGGGUUGCC (((....)))..((((((((....(((...(((((((((((((((...(((((((....))))))).)))).)))).))...))))).....))).....))))))))(((......))) ( -53.00) >DroYak_CAF1 73974 87 - 1 GUCCGAAGGCGACAAGUGUUAGUGCCCACUUCGGACCACGCCCACAUGA----CCAAGUGGGCG-----------------------------GGAGUGGAGGACAUGGGCAGGGUUGCC .......((((((.........(((((((((((..((.(((((((....----....)))))))-----------------------------))..)))))....))))))..)))))) ( -37.00) >consensus AUCUGAAGGCGACAAGUGUUAGUGCCCAAAUCGGACCACGCCCACAUGC____CCAAGUGGGCG______________________GGUAAAGGGAGUUGAGGACAUGGGCAGAGUUGCC .......((((((.........((((((..........(((((((.((......)).)))))))................................((.....)).))))))..)))))) (-28.06 = -27.94 + -0.12)



| Location | 11,337,573 – 11,337,673 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -41.54 |
| Consensus MFE | -28.62 |
| Energy contribution | -29.94 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11337573 100 + 27905053 ----------------CGCCCACUUGG----GCAUGUGGGCGUGGUCCGAUUUGGGCACUAACACUUGUCGCCUUCAGAUGACCGAUUGCAACGUGGCUUUUGGACAGAGAUCGCUCAGG ----------------.((((((....----....))))))(((((((.((((((((((........)).)))..)))))..((((..((......))..))))...).))))))..... ( -33.60) >DroSec_CAF1 76397 100 + 1 ----------------CGCCCACUUGG----GCAUGUGGGCGUGGUCCGAUUUAGGCACUAACACUUGUCGCCUUCAGAUGACCGAUUGCGACGGGGUUGCUAGAUGGAGAUCGCUCAUG ----------------.((((((....----....))))))(((((((.((((((....((((.((((((((..((........))..)))))))))))))))))).).))))))..... ( -41.50) >DroSim_CAF1 74995 100 + 1 ----------------CGCCCACUUGG----GCAUGUGGGCGUGGUCCGAUUUGGGCCCUAACACUUGUCGCCUUCAGAUGACCGAUUGCGACGGUGUUGCUAGAUAGAGAUCGCUCAUG ----------------.((((....))----))..(((((((.(((((.....))))).(((((((.(((((..((........))..))))))))))))............))))))). ( -40.40) >DroEre_CAF1 72292 120 + 1 ACCUACACUCCACUGCCACCCACUUGGGCGGCCAUGUGGGCGUGGUCCGAUUUGGGCACUAACACUUGUCGCCCUUGGACGACCGAUGGCGACGAGCAUGUUGGACAGAGAUCGCUCCUG .....((((((((.(((.(((....))).)))...))))).)))((((.....)))).(((((((((((((((.((((....)))).)))))))))..)))))).(((.((....))))) ( -52.70) >DroYak_CAF1 74001 100 + 1 ----------------CGCCCACUUGG----UCAUGUGGGCGUGGUCCGAAGUGGGCACUAACACUUGUCGCCUUCGGACGAACGCUGGCGAAGAGCAUGUUGCACAGAGAACACUCCUG ----------------.((((((....----....))))))(((((((((((..((((........))))..))))))))...(.(((((((........)))).))).)..)))..... ( -39.50) >consensus ________________CGCCCACUUGG____GCAUGUGGGCGUGGUCCGAUUUGGGCACUAACACUUGUCGCCUUCAGAUGACCGAUUGCGACGAGGAUGUUGGACAGAGAUCGCUCAUG .................((((((.((......)).))))))((((((...((((....((((((((((((((..((........))..))))))))..)))))).))))))))))..... (-28.62 = -29.94 + 1.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:32 2006