| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,332,569 – 11,332,963 |
| Length | 394 |
| Max. P | 0.985191 |

| Location | 11,332,569 – 11,332,682 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332569 113 + 27905053 AGUCUGCGGAUACUUUUGCAGCGUCAUCGCAUGAUUACGUCGAUUUUGGGACCCAGUAAUCAUCUGGUGCACGAACACAUAUGUAC-------ACCCCAAUCUCAGCUUCCGCAUCCCAU .(..((((((..((......((((.(((....))).)))).((..(((((...(((.......)))(((((..........)))))-------..)))))..))))..))))))..)... ( -28.80) >DroSec_CAF1 71777 113 + 1 AGUCUGCGGAUACUUUUGCAGCGUCAUCGCAUGAUUACGUCGAUUUUGGGACCCAGUAAUCAUCUGGUGCACGAACACAUAUGUAC-------ACCCCAAUCUCAGCUUCCGCAUCCCAU .(..((((((..((......((((.(((....))).)))).((..(((((...(((.......)))(((((..........)))))-------..)))))..))))..))))))..)... ( -28.80) >DroSim_CAF1 69989 113 + 1 AGUCUGCGGAUACUUUUGCAGCGUCAUCGCAUGAUUACGUCGAUUUUGGGACCCAGUAAUCAUCUGGUGCACGAACACAUAUGUAC-------ACCCCAAUCUCAGCUUCCGCAUCCCAU .(..((((((..((......((((.(((....))).)))).((..(((((...(((.......)))(((((..........)))))-------..)))))..))))..))))))..)... ( -28.80) >DroEre_CAF1 67259 112 + 1 AGUCUGCGGAUACUUUUGCAGCGUCAUCGCAUGAUUACGUCGAUUGUGGGACCCAGUAAUCAUCUGGUGCACGAACACAUGUGUAC-------A-CCCAAUCUCAGCUUCCGCAUCCCAU .(..((((((..((......((((.(((....))).)))).(((((.((....(((.......)))(((((((......)))))))-------.-)))))))..))..))))))..)... ( -31.80) >DroYak_CAF1 69029 120 + 1 AGUCUGCGGAUACUUUUGCAGCGUCAUCGCAUGAUUACGUCGAUUUUGGGACCCAGUAAUCAUCUGGUGCACGAACACAUAUGUACUAUGUACACCCCAAUCCCAGCUUCCGCAUCCCAU .(..((((((..((.......((((((((.(((((((((((........)))...)))))))).))))).)))........(((((...)))))..........))..))))))..)... ( -30.70) >consensus AGUCUGCGGAUACUUUUGCAGCGUCAUCGCAUGAUUACGUCGAUUUUGGGACCCAGUAAUCAUCUGGUGCACGAACACAUAUGUAC_______ACCCCAAUCUCAGCUUCCGCAUCCCAU .(..((((((..((.......((((((((.(((((((((((........)))...)))))))).))))).))).(((....)))....................))..))))))..)... (-27.88 = -27.88 + 0.00)

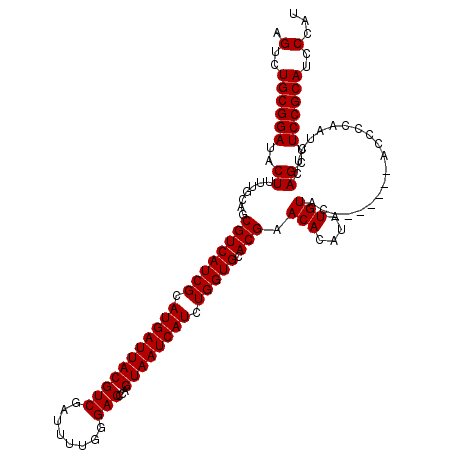

| Location | 11,332,569 – 11,332,682 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.20 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -33.60 |
| Energy contribution | -33.84 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332569 113 - 27905053 AUGGGAUGCGGAAGCUGAGAUUGGGGU-------GUACAUAUGUGUUCGUGCACCAGAUGAUUACUGGGUCCCAAAAUCGACGUAAUCAUGCGAUGACGCUGCAAAAGUAUCCGCAGACU ..((..((((((.(((....(((.(((-------((.(((..((((....))))...(((((((((.(((......))).).))))))))...)))))))).))).))).))))))..)) ( -37.30) >DroSec_CAF1 71777 113 - 1 AUGGGAUGCGGAAGCUGAGAUUGGGGU-------GUACAUAUGUGUUCGUGCACCAGAUGAUUACUGGGUCCCAAAAUCGACGUAAUCAUGCGAUGACGCUGCAAAAGUAUCCGCAGACU ..((..((((((.(((....(((.(((-------((.(((..((((....))))...(((((((((.(((......))).).))))))))...)))))))).))).))).))))))..)) ( -37.30) >DroSim_CAF1 69989 113 - 1 AUGGGAUGCGGAAGCUGAGAUUGGGGU-------GUACAUAUGUGUUCGUGCACCAGAUGAUUACUGGGUCCCAAAAUCGACGUAAUCAUGCGAUGACGCUGCAAAAGUAUCCGCAGACU ..((..((((((.(((....(((.(((-------((.(((..((((....))))...(((((((((.(((......))).).))))))))...)))))))).))).))).))))))..)) ( -37.30) >DroEre_CAF1 67259 112 - 1 AUGGGAUGCGGAAGCUGAGAUUGGG-U-------GUACACAUGUGUUCGUGCACCAGAUGAUUACUGGGUCCCACAAUCGACGUAAUCAUGCGAUGACGCUGCAAAAGUAUCCGCAGACU ..((..((((((.(((....(((((-(-------((((((....))..)))))))..(((((((((.(((......))).).))))))))(((....)))..))).))).))))))..)) ( -36.60) >DroYak_CAF1 69029 120 - 1 AUGGGAUGCGGAAGCUGGGAUUGGGGUGUACAUAGUACAUAUGUGUUCGUGCACCAGAUGAUUACUGGGUCCCAAAAUCGACGUAAUCAUGCGAUGACGCUGCAAAAGUAUCCGCAGACU ..((..((((((.(((....(((.(((((.(((.((......((((....))))...(((((((((.(((......))).).)))))))))).)))))))).))).))).))))))..)) ( -38.50) >consensus AUGGGAUGCGGAAGCUGAGAUUGGGGU_______GUACAUAUGUGUUCGUGCACCAGAUGAUUACUGGGUCCCAAAAUCGACGUAAUCAUGCGAUGACGCUGCAAAAGUAUCCGCAGACU ..((..((((((.(((((..((((((........((((..........)))).((((.......)))).))))))..))(.(((.(((....))).))))......))).))))))..)) (-33.60 = -33.84 + 0.24)


| Location | 11,332,649 – 11,332,748 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.62 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332649 99 - 27905053 AAUCCGCUCCUUGUACUCCUGCCCCAUCUCCUGCUUC------------UGCUCCUGGUCUAGUCG--UUCGUCUGGGUAAUGGGAUGCGGAAGCUGAGAUUGGGGU-------GUACAU ...........(((((....((((((((((..(((((------------((((((((.(((((.(.--...).)))))...))))).)))))))).)))).))))))-------))))). ( -43.70) >DroSec_CAF1 71857 113 - 1 AAUCCGCUCCUGCCACUCCUGCUCCACCUCCUGCUUCUGCUUCUGCCUCUGCUCCUGGUCUAGUCGAGUUCGUCUGGGUAAUGGGAUGCGGAAGCUGAGAUUGGGGU-------GUACAU .....((....)).......((((((.(((..((((((((....((....))(((((.(((((.((....)).)))))...))))).)))))))).)))..))))))-------...... ( -36.10) >DroSim_CAF1 70069 113 - 1 AAUCCGCUCCUGCCACUCCUGCUCCACCUCCUGCUUCUGCUUCUGCUUCUGCUCCUGGUCUAGUCGAUUUCGUCUGGGUAAUGGGAUGCGGAAGCUGAGAUUGGGGU-------GUACAU .....((....))((((((..(((........((....))....(((((((((((((.(((((.((....)).)))))...))))).)))))))).)))...)))))-------)..... ( -40.20) >DroEre_CAF1 67339 88 - 1 AAUCCAAUCC----------------GCUCCUGCUCC------UGCUCCUGCUCCUGGUCUAGUCG--UUCGUCUGGGUAAUGGGAUGCGGAAGCUGAGAUUGGG-U-------GUACAC .((((((((.----------------((....))((.------.(((.(((((((((.(((((.(.--...).)))))...))))).)))).))).)))))))))-)-------...... ( -30.30) >DroYak_CAF1 69109 109 - 1 AAUCCGCUCCUG---UUCCUGCUUCACCUCCUGCUUC------ACCUCCUGCCCCUGGUCUAUUCG--CUCGUCUGGGUAAUGGGAUGCGGAAGCUGGGAUUGGGGUGUACAUAGUACAU ....((((((.(---((((.(((((......(((.((------.((...((((((.(((......)--)).)...)))))..)))).)))))))).))))).))))))............ ( -33.30) >consensus AAUCCGCUCCUG__ACUCCUGCUCCACCUCCUGCUUC______UGCUCCUGCUCCUGGUCUAGUCG__UUCGUCUGGGUAAUGGGAUGCGGAAGCUGAGAUUGGGGU_______GUACAU ....................(((((..(((..(((((............((((((((.(((((.((....)).)))))...))))).)))))))).)))...)))))............. (-20.57 = -21.62 + 1.05)



| Location | 11,332,748 – 11,332,866 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.29 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -35.78 |
| Energy contribution | -35.22 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332748 118 + 27905053 GGGUGCUUAGACCGGACACAAUUAUCGGGGGCCAAAAGGGCUUUCAAUUUCACUUGGUUAGCUGCAAGUAAACAGCUUUGCGGCUCCACGACUUCAGCUGGCU--UUGGGUGGCGCUCAA (((((((...(((.............(..((((.....))))..).......(..(((((((((.((((....((((....)))).....)))))))))))))--..))))))))))).. ( -43.20) >DroSec_CAF1 71970 118 + 1 GGGUGCUCAGACCGGACACAAUUAUCGUGGGCCAAAAGGGCUUUCAAUUUCACUUGGUUAGCUGCAAGUAAACAGCUUUGCGGCCCCACGACUUCAGCUGGCU--UUGGGUGGCGCUCAA (((((((((((((((.........(((((((((.....((((.((((......))))..))))((((((.....)).)))))).)))))))......)))).)--)))...))))))).. ( -40.86) >DroSim_CAF1 70182 118 + 1 GGGUGCUCAGACCGGACACAAUUAUCGUGGGCCAAGAGGGCUUUCAAUUUCACUUGGUUAGCUGCAAGUAAACAGCUUUGCGGCCCCACGACUUCAGCUGGCU--UUGGGUGGCGCUCAA (((((((((((((((.........((((((((((((.(((........))).)))))...(((((((((.....)).))))))))))))))......)))).)--)))...))))))).. ( -43.36) >DroEre_CAF1 67427 118 + 1 GGGUGCUCAGACCGGACACAAUUAUCGGGGACCAAGAGGGCUUUCAAUUUCACUUGGUUAGCUGCAAGUAAACAGCUUUGCGGCUCCUCGAUUUCAGCUGGCU--UUGGGUGGCGCUCAA (((((((((((((((........(((((((((((((.(((........))).)))))).((((((((((.....)).))))))))))))))).....)))).)--)))...))))))).. ( -44.12) >DroYak_CAF1 69218 120 + 1 GGGUGCUCUGACCGGACACAAUUAUCGGGGACCAAAAGGGCUUUCAAUUUCACUUGGUUAGCUGCAAGUAAACAGCUUUGCGGCUCCUCGAUUUCAGCCGGCUUUUUGGGUGGCGCUCAA ((((((((..(((((........((((((((((((..(((........)))..))))).((((((((((.....)).))))))))))))))).....))))....)..)..))))))).. ( -40.12) >consensus GGGUGCUCAGACCGGACACAAUUAUCGGGGGCCAAAAGGGCUUUCAAUUUCACUUGGUUAGCUGCAAGUAAACAGCUUUGCGGCUCCACGACUUCAGCUGGCU__UUGGGUGGCGCUCAA (((((((.((.((((.........(((((((((.....((((.((((......))))..))))((((((.....)).)))))).)))))))......))))))........))))))).. (-35.78 = -35.22 + -0.56)

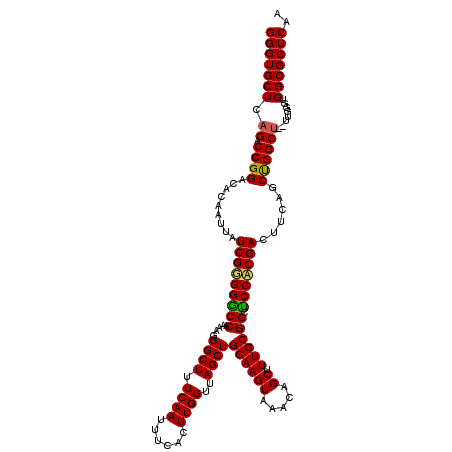

| Location | 11,332,828 – 11,332,939 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.24 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -34.86 |
| Energy contribution | -34.30 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332828 111 + 27905053 CGGCUCCACGACUUCAGCUGGCU--UUGGGUGGCGCUCAAGUCGAGCAUGCAACCAUCACAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUC ..((((...(((((.(((..(((--......)))))).)))))))))......(((((...((((((((((....)).(-------((......)))..)))))..)))...)))))... ( -35.30) >DroSec_CAF1 72050 111 + 1 CGGCCCCACGACUUCAGCUGGCU--UUGGGUGGCGCUCAAGUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUC .((((...(((....(((((((.--.(((...((((((.....))))..))..)))..))....(((((((....)).(-------((......)))..)))))))))).)))..)))). ( -37.00) >DroSim_CAF1 70262 111 + 1 CGGCCCCACGACUUCAGCUGGCU--UUGGGUGGCGCUCAAGUCGGGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUC ..((((...(((((.(((..(((--......)))))).)))))))))......((((((..((((((((((....)).(-------((......)))..)))))..)))..))))))... ( -40.00) >DroEre_CAF1 67507 118 + 1 CGGCUCCUCGAUUUCAGCUGGCU--UUGGGUGGCGCUCAAGUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAUCAAGCAUCAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUC ..((..((((((((.(((..(((--......)))))).))))))))...))..((((((..((((((((((....))........(((......)))..)))))..)))..))))))... ( -39.60) >DroYak_CAF1 69298 113 + 1 CGGCUCCUCGAUUUCAGCCGGCUUUUUGGGUGGCGCUCAAGUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUC ..((..((((((((.((((.(((.....))).).))).))))))))...))..((((((..((((((((((....)).(-------((......)))..)))))..)))..))))))... ( -41.60) >consensus CGGCUCCACGACUUCAGCUGGCU__UUGGGUGGCGCUCAAGUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU_______CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUC .((((...(((....((((((((.....(((.((((((.....))))..)).)))......)))(((((((....)).........((......))...)))))))))).)))..)))). (-34.86 = -34.30 + -0.56)


| Location | 11,332,866 – 11,332,963 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -28.44 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332866 97 + 27905053 GUCGAGCAUGCAACCAUCACAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUCU----------------CCAGGACUCCCGACUCCGGCUCA ...((((......(((((...((((((((((....)).(-------((......)))..)))))..)))...)))))....----------------...(((........))).)))). ( -28.60) >DroSec_CAF1 72088 97 + 1 GUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUCU----------------CCAGGACUCCCGACUCCGGCUCA ...((((......((((((..((((((((((....)).(-------((......)))..)))))..)))..))))))....----------------...(((........))).)))). ( -31.20) >DroSim_CAF1 70300 97 + 1 GUCGGGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUCU----------------CCAGGACUCCCGACUCCGGCUCA ((((((..((...((((((..((((((((((....)).(-------((......)))..)))))..)))..))))))....----------------.)).....))))))......... ( -31.40) >DroEre_CAF1 67545 104 + 1 GUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAUCAAGCAUCAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUCU----------------CCAGGACUCCCCACUCCGGCUCA ...((((......((((((..((((((((((....))........(((......)))..)))))..)))..))))))....----------------...(((........))).)))). ( -31.20) >DroYak_CAF1 69338 113 + 1 GUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU-------CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUCUCGAGGACUCCAGGACUCCAGGACUCCCGACUCCGGCUCA ...((((......((((((..((((((((((....)).(-------((......)))..)))))..)))..))))))...(((.(((.(((.((...)).))).)))))).....)))). ( -39.60) >consensus GUCGAGCAUGCAACCAUCGCAAGCGGCACGCAAACGCAU_______CAAAUCAAUGAAAGUGCCCGGCUAUCGAUGGCUCU________________CCAGGACUCCCGACUCCGGCUCA ...((((......((((((..((((((((((....)).........((......))...)))))..)))..)))))).......................(((........))).)))). (-28.44 = -28.48 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:24 2006