
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,331,748 – 11,332,132 |
| Length | 384 |
| Max. P | 0.999441 |

| Location | 11,331,748 – 11,331,853 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.84 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.63 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.533925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331748 105 - 27905053 AUAAUGAAAUUCCUAGCUGCUUUGUGCCCACGACGGGCACAAGCCACUUGGCGGGAUGUCCCGAUCCCUGAUCCAUCCCCUACAUAUAUAUUCAUAUAUAUGUAU ...........((.((..((((.((((((.....))))))))))..)).)).((((((....((((...)))))))))).((((((((((....)))))))))). ( -36.10) >DroEre_CAF1 66499 83 - 1 AUAAUGAAAUUCCUAGCUGCUUUGUGCCCACGACG-----AAGCCACUUGGCGGGAUGUCCCGAUCCCUGAUCUGUCCCCUACAUAUA----------------- ...(((.....((.((..(((((((.......)))-----))))..)).)).((((((....((((...))))))))))...)))...----------------- ( -17.70) >DroYak_CAF1 68223 93 - 1 AUAAUGAAAUUCCUAGCUGCUUUGUGCCCACGACG-----AAGCCACUUGGCGGGAUGUCCCGAUCCCUGAUCCGUCGCCUACAUAUAUACACAUAUA------- ......................((((....(((((-----..(((....)))(((((......))))).....)))))....))))............------- ( -20.40) >consensus AUAAUGAAAUUCCUAGCUGCUUUGUGCCCACGACG_____AAGCCACUUGGCGGGAUGUCCCGAUCCCUGAUCCGUCCCCUACAUAUAUA__CAUAUA_______ ...(((.........((........))....((((.......(((....)))(((((......))))).....)))).....))).................... (-15.52 = -15.63 + 0.11)

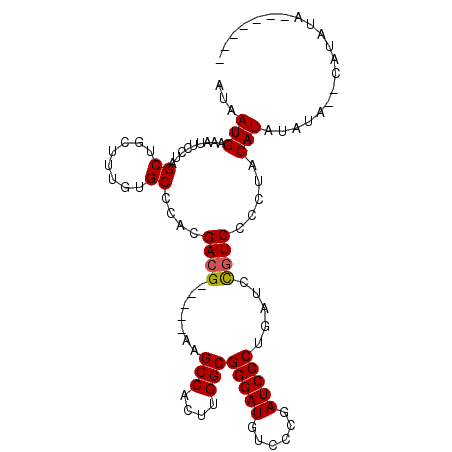

| Location | 11,331,853 – 11,331,972 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -28.52 |
| Energy contribution | -29.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331853 119 + 27905053 AUGCACCCCGGGCCAAAGGACAAAGACAUGUUUUUAAUAGUUUCGCAUGUUUGUGCAGCAGGCUUUGUUUUUUUUUUUUUUUUUGGCCCCUCCUU-GGAGGAGGCUGCAGGAACCCCUCU .((((.(((((((((((((((((((...((((............((((....))))))))..)))))))...........)))))))))(((...-.)))).)).))))((....))... ( -34.20) >DroSec_CAF1 71037 112 + 1 AUGCACCCCGGGCCAAAGGACAAAGACAUGUUUUUAAUAGUUUCGCAUGUUUGUGCAGCAGGGUUUGUUUUU-------UUUUUGGCCCCUCCUU-GGAGGAUUCUGCAUGAUCCCCCCU (((((....(((((((((((..((((((..((((.....(((.((((....)))).)))))))..)))))))-------))))))))))(((...-.))).....))))).......... ( -31.30) >DroSim_CAF1 69252 109 + 1 AUGCACCCCGGGCCAAAGGACAAAGACAUGUUUUUAAUAGUUUCGCAUGUUUGUGCAGCAGGCUUUGUUUUU--------UUUUGGCCCCUCC---GGAGGAGGCUGCAUGAUCCCCUCU (((((((((((((((((((((((((...((((............((((....))))))))..)))))))...--------)))))))))(...---.).)).)).))))).......... ( -33.30) >DroEre_CAF1 66582 110 + 1 AUGCACCCCGGGCCAAAGGACAAAGACAUGUUUUUAAUAGUUUCGCAUGUUUGUGCAGCAGGCUUUGUUUU---------UUUUGGCCCCUCCUC-GGAGGAGGCUGGAGGAUCCUGGCC .(((((.((........))....((((((((.............)))))))))))))...((((..(..((---------(((..(((((((...-.)))).)))..)))))..).)))) ( -39.32) >DroYak_CAF1 68316 113 + 1 AUGCACCCCGGGCCAAAGGACAAAGACAUGUUUUUAAUAGUUUCGCAUGUUUGUGCAGCAGGCUUUGUUUUU-------UUUUUGGCCCCUCCUCUGGAGGAGGCUGGAGGAUCCCCAGU .........((((((((((((((((...((((............((((....))))))))..)))))))...-------.)))))))))(((((....)))))(((((.(....)))))) ( -38.40) >consensus AUGCACCCCGGGCCAAAGGACAAAGACAUGUUUUUAAUAGUUUCGCAUGUUUGUGCAGCAGGCUUUGUUUUU_______UUUUUGGCCCCUCCUC_GGAGGAGGCUGCAGGAUCCCCUCU .((((.((.((((((((((((((((...((((............((((....))))))))..)))))))...........)))))))))((((...))))..)).))))........... (-28.52 = -29.52 + 1.00)

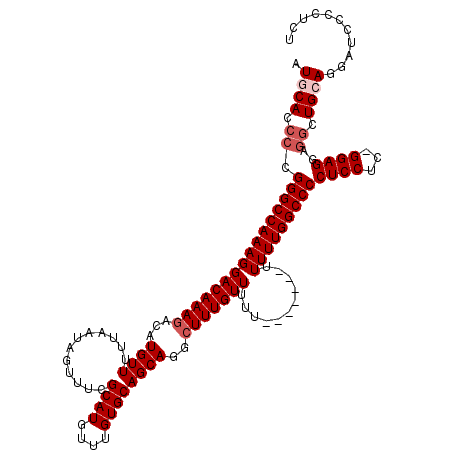

| Location | 11,331,972 – 11,332,092 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.16 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -34.88 |
| Energy contribution | -34.88 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331972 120 + 27905053 UCCCCUUUUAGUUUCCCCUUUCAGUUCCGAUUGUGUGCAGUAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAA ..........................((((((.(((((.....))))).)))))).......((((((((((((((.(((((.(((.((....))))).))))).)))))).)))))))) ( -35.50) >DroSec_CAF1 71149 120 + 1 UCCCGUUUUAGUUUCCCCUUUCAGUUCCGAUUGUGUGCAGUAUGCGCAAAGUUGGUCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAA ..........................((((((.(((((.....))))).)))))).......((((((((((((((.(((((.(((.((....))))).))))).)))))).)))))))) ( -35.70) >DroSim_CAF1 69361 120 + 1 UCACCUUUUAGUUUCCUCUUUCAGUUCCGAUUGUGUGCAGUAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAA ..........................((((((.(((((.....))))).)))))).......((((((((((((((.(((((.(((.((....))))).))))).)))))).)))))))) ( -35.50) >DroEre_CAF1 66692 118 + 1 CCC--UAUCCCUUUCCCCUCGCAGUUCCGAUUGUGUGCAGUAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUCGUGCAGAGCAUAAA ...--.....................((((((.(((((.....))))).)))))).......(((((((((((((..(((((.(((.((....))))).)))))..))))).)))))))) ( -34.10) >DroYak_CAF1 68429 115 + 1 UCC--UA---GUUUCCCCUUUCAGUUCCGAUUGUGUGCAGUGCGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUCUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAA ...--..---................((((((.(((((.....))))).)))))).......((((((((((((((.(((((.(((.((....))))).))))).)))))).)))))))) ( -35.20) >consensus UCCC_UUUUAGUUUCCCCUUUCAGUUCCGAUUGUGUGCAGUAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAA ..........................((((((.(((((.....))))).)))))).......(((((((((((((..(((((.(((.((....))))).)))))..))))).)))))))) (-34.88 = -34.88 + -0.00)



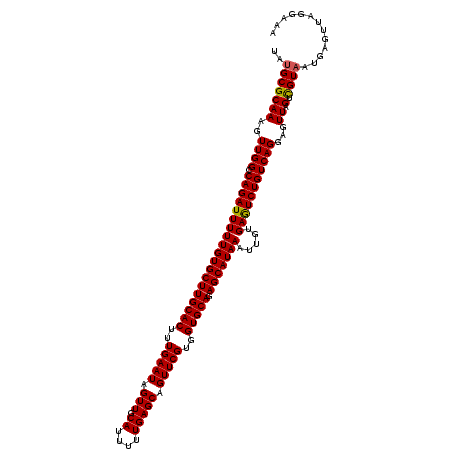
| Location | 11,332,012 – 11,332,132 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -31.72 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11332012 120 + 27905053 UAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAAUUGUGAGUCUGUCAGGAGUUAGUUGUAAUGAGUAAGGAAA .....((((..(((((.(((((((((((((((((((.(((((.(((.((....))))).))))).)))))).))))))).....))))))))))).......)))).............. ( -31.10) >DroSec_CAF1 71189 120 + 1 UAUGCGCAAAGUUGGUCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAAUUGUGAGUCUGUCAGUAGUUAGUCGUAAUGAGUUAGGGAA ..(((((.((.(..(.((((((((((((((((((((.(((((.(((.((....))))).))))).)))))).))))))).....))))))).)...).)).).))))............. ( -33.20) >DroSim_CAF1 69401 120 + 1 UAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAAUUGUGAGUCUGUCAGGAGUUAGUCGUAAUGAGUUAGGAAA ..(((((((..(((((.(((((((((((((((((((.(((((.(((.((....))))).))))).)))))).))))))).....)))))))))))...)).).))))............. ( -33.00) >DroEre_CAF1 66730 120 + 1 UAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUCGUGCAGAGCAUAAAUUGUGAAUCUGUCAGGAGUUAGUCGUAAUGAGUUGGGAAA ..(((((((..(((((.((((((((((((((((((..(((((.(((.((....))))).)))))..))))).))))))).....)))))))))))...)).).))))............. ( -31.60) >DroYak_CAF1 68464 120 + 1 UGCGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUCUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAAUUGUGAGUCUGUCAGGAGUUGGUCGUAAUGAGUUGGGGAA (((((.(((..(((((.(((((((((((((((((((.(((((.(((.((....))))).))))).)))))).))))))).....)))))))))))...)))).))))............. ( -36.00) >consensus UAUGCGCAAAGUUGGCCAGAUUUUUGUGCUUGCACUUUGAAUAGUUGCAUUUUUGAGCAGUUCGUGGUGCAGAGCAUAAAUUGUGAGUCUGUCAGGAGUUAGUCGUAAUGAGUUAGGAAA ..(((((((..((((.(((((((((((((((((((..(((((.(((.((....))))).)))))..))))).))))))).....)))))))))))...)).).))))............. (-31.72 = -31.60 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:17 2006