| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,331,326 – 11,331,509 |
| Length | 183 |
| Max. P | 0.999872 |

| Location | 11,331,326 – 11,331,438 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -25.98 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331326 112 + 27905053 UUUUAAAUUUGCACAAAAUUGCAAAUAACUUGCAAUCAAGGUGCAAGAAGUACGCAAAAGGGCUGGGGU--------UGGGAAUGGGGUUCCAGUGACUUUGGAGUCACAACAAAAUGGC .......(((((((...(((((((.....)))))))....)))))))..............((((...(--------(((((((...))))).((((((....))))))..)))..)))) ( -30.20) >DroSec_CAF1 70503 112 + 1 UUUUAAAUUUGCACAAAAUUGCAAAUAACUUGCAAUCAAGGUGCAAGAAGUACGCAAAAGGGCUGGGGU--------UGGGAAUGGGGUUCCAGUGACUUUGGAGUCACAACAAAAUGGC .......(((((((...(((((((.....)))))))....)))))))..............((((...(--------(((((((...))))).((((((....))))))..)))..)))) ( -30.20) >DroSim_CAF1 68722 112 + 1 UUUUAAAUUUGCACAAAAUUGCAAAUAACUUGCAAUCAAGGUGCAAGAAGUACGCAAAAGGGCUGGGGU--------UGGGAAUGGGGUUCCAGUGACUUUGGAGUCACAACAAAAUGGC .......(((((((...(((((((.....)))))))....)))))))..............((((...(--------(((((((...))))).((((((....))))))..)))..)))) ( -30.20) >DroEre_CAF1 66074 105 + 1 UUUUAAAUUUGCACAAAAUUGCAAAUAACUUGCAAUCAAGGUGCAAGAAGUACGGAAAAGGGCUGG---------------GAUGGGGUUCCAGUGACUCUGGAGUCACAACAAAAUGGC .......(((((((...(((((((.....)))))))....))))))).((..(......)..)).(---------------((......))).((((((....))))))........... ( -27.90) >DroYak_CAF1 67785 120 + 1 UUUUAAAUUUGCACAAAAUUGCAAAUAACUUGCAAUCAAGGUGCAAGAAGUGCGGCAAAGGGCUGGGAAUGCAUCCAUGGGGAUGGGGUUCCAGUGACUUUGGAGUCACAACAAAAUGGC .......(((((((...(((((((.....)))))))....)))))))..((((..(((((.(((((((.(.(((((....))))).).)))))))..)))))..).)))........... ( -42.00) >consensus UUUUAAAUUUGCACAAAAUUGCAAAUAACUUGCAAUCAAGGUGCAAGAAGUACGCAAAAGGGCUGGGGU________UGGGAAUGGGGUUCCAGUGACUUUGGAGUCACAACAAAAUGGC .......(((((((...(((((((.....)))))))....)))))))..............(((...............(((((...))))).((((((....))))))........))) (-25.98 = -26.14 + 0.16)

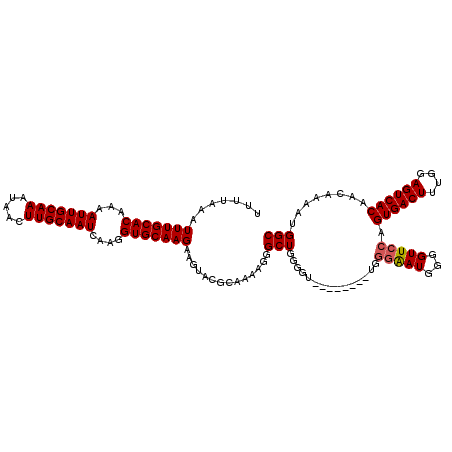
| Location | 11,331,326 – 11,331,438 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.33 |
| SVM RNA-class probability | 0.999872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331326 112 - 27905053 GCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCCCA--------ACCCCAGCCCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAAUUUUGUGCAAAUUUAAAA ((......(((((((((....)))))).((((.....))))..--------.....)))......)).......((((((...(((((((((...)))))))))..))))))........ ( -25.10) >DroSec_CAF1 70503 112 - 1 GCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCCCA--------ACCCCAGCCCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAAUUUUGUGCAAAUUUAAAA ((......(((((((((....)))))).((((.....))))..--------.....)))......)).......((((((...(((((((((...)))))))))..))))))........ ( -25.10) >DroSim_CAF1 68722 112 - 1 GCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCCCA--------ACCCCAGCCCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAAUUUUGUGCAAAUUUAAAA ((......(((((((((....)))))).((((.....))))..--------.....)))......)).......((((((...(((((((((...)))))))))..))))))........ ( -25.10) >DroEre_CAF1 66074 105 - 1 GCCAUUUUGUUGUGACUCCAGAGUCACUGGAACCCCAUC---------------CCAGCCCUUUUCCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAAUUUUGUGCAAAUUUAAAA ((.........((((((....))))))(((....)))..---------------...))...............((((((...(((((((((...)))))))))..))))))........ ( -24.90) >DroYak_CAF1 67785 120 - 1 GCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUCCCCAUGGAUGCAUUCCCAGCCCUUUGCCGCACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAAUUUUGUGCAAAUUUAAAA ((.........((((((....)))))).((((...(((((....)))))..))))............)).....((((((...(((((((((...)))))))))..))))))........ ( -31.90) >consensus GCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUCCCA________ACCCCAGCCCUUUUGCGUACUUCUUGCACCUUGAUUGCAAGUUAUUUGCAAUUUUGUGCAAAUUUAAAA ((.........((((((....))))))(((....)))....................))...............((((((...(((((((((...)))))))))..))))))........ (-24.10 = -24.10 + -0.00)

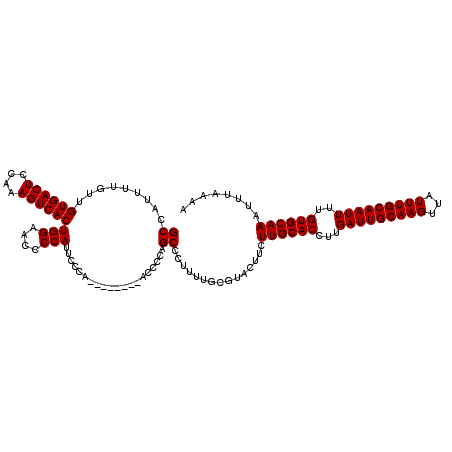
| Location | 11,331,398 – 11,331,509 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.29 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -22.72 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
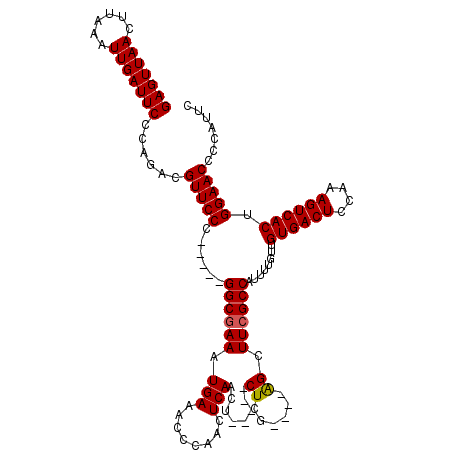
>3R_DroMel_CAF1 11331398 111 - 27905053 GAGUUAACUUAAAUUGAUUCCCAGACGUUCCC-----GGCGAAAUGAAACCCAACUCAACUCAACGCUCG----AGCUUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUC (((((((......)))))))......(((((.-----((((((.(((........))).(((.......)----)).))))))........((((((....)))))).)))))....... ( -27.20) >DroSec_CAF1 70575 106 - 1 GAGUUAACUUAAAUUGAUUCCCAGACGUUCCC-----GGCAAAAUGAAACCCAACUCAACU-----CUCG----AGCUUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUC (((((((......)))))))......(((((.-----(((((((((........(((....-----...)----))......)))))))))((((((....)))))).)))))....... ( -23.74) >DroSim_CAF1 68794 106 - 1 GAGUUAACUUAAAUUGAUUCCCAGACGUUCCC-----GGCGAAAUGAAACCCAACUCAACU-----CUCG----AGCUUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUC (((((((......)))))))......(((((.-----((((((.((.....)).(((....-----...)----)).))))))........((((((....)))))).)))))....... ( -25.80) >DroEre_CAF1 66140 108 - 1 GAGUUAACUUAAAUUGAUUCCCAGACGUUCCC-----GGCGAAAUGAAACCCAACUCAA-U-----CUCGGGCGGGCUUCGCCAUUUUGUUGUGACUCCAGAGUCACUGGAACCCCAUC- (((((((......)))))))......(((((.-----((((((......(((..(((..-.-----...))).))).))))))........((((((....)))))).)))))......- ( -30.00) >DroYak_CAF1 67865 110 - 1 GAGUUAACUUAAAUUGAUUCCCAGACGUUCCCGGCCCGGCGAAAUGAAACCCAACUCAA-U-----CUCG----GGCUUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUCC (((((((......)))))))......(((((.(((((((.((..(((........))).-)-----))))----)))).............((((((....)))))).)))))....... ( -28.10) >consensus GAGUUAACUUAAAUUGAUUCCCAGACGUUCCC_____GGCGAAAUGAAACCCAACUCAACU_____CUCG____AGCUUCGCCAUUUUGUUGUGACUCCAAAGUCACUGGAACCCCAUUC (((((((......)))))))......(((((......((((((.(((........)))........((......)).))))))........((((((....)))))).)))))....... (-22.72 = -22.68 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:13 2006