| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
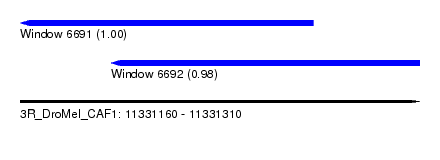
| Location | 11,331,160 – 11,331,310 |
| Length | 150 |
| Max. P | 0.998995 |

| Location | 11,331,160 – 11,331,270 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -26.96 |
| Energy contribution | -26.48 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331160 110 - 27905053 AU----CCACCCACACACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUUUUUUAUGCAAUUUGUGCCGGAAAAGCAGGAAUA------C .(----((............((((.........)))).((((((((.(((((((((.....)))))))))(((..((....))..))).....))))))))......)))...------. ( -26.80) >DroSec_CAF1 70333 114 - 1 AUCCAUCCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUUUUUUAUGCAAUUUGUGCCGGAAAAGCAGGAAUA------C .....(((............((((.........)))).((((((((.(((((((((.....)))))))))(((..((....))..))).....))))))))......)))...------. ( -26.80) >DroSim_CAF1 68552 114 - 1 AUCCAUCCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUUUUUUAUGCAAUUUGUGCCGGAAAAGCAGGAAUA------C .....(((............((((.........)))).((((((((.(((((((((.....)))))))))(((..((....))..))).....))))))))......)))...------. ( -26.80) >DroEre_CAF1 65892 116 - 1 AU----CCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGCGCGCGAAAUUUUUCAUGCAAUUUGUGCCGGAAAAGCAGGAAUACCCGCAC .(----((............((((.........))))....(((((.(((((((((.....)))))))))(((.(((....))).))).....))))))))...((.((....)).)).. ( -32.20) >DroYak_CAF1 67593 112 - 1 AU----CCAC----CCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGCGCGCGAAAUUUUUCAUGCAAUUUGUGCCGGAAAAGCAGGAAUACUCGUGC .(----((..----......((((.........))))....(((((.(((((((((.....)))))))))(((.(((....))).))).....))))))))...(((.((....)).))) ( -32.50) >consensus AU____CCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUUUUUUAUGCAAUUUGUGCCGGAAAAGCAGGAAUA______C .................(((((((.........)))).((((((((.(((((((((.....)))))))))(((.(((....))).))).....)))))))).....)))........... (-26.96 = -26.48 + -0.48)



| Location | 11,331,194 – 11,331,310 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.76 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -26.07 |
| Energy contribution | -26.03 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11331194 116 - 27905053 GGCGAAGGAAAAUGAAGGAAAAUCCAGCCAGACGACCACCAU----CCACCCACACACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUU ((((((((....((((((....((......)).((......)----)..........))))))))))).)))......(((.((.(.(((((((((.....)))))))))).)).))).. ( -28.00) >DroSec_CAF1 70367 120 - 1 UGCGAAGGAAAAUGAAGGAAAAUCCAGCCAGACGACCACCAUCCAUCCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUU .(((((((....((((((....((......)).((......))..............))))))))))).(((((.....)))))...(((((((((.....)))))))))..))...... ( -27.90) >DroSim_CAF1 68586 120 - 1 GGCGAAGGAAAAUGAAGGAAAAUCCAGCCAGACGACCACCAUCCAUCCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUU ((((((((....((((((....((......)).((......))..............))))))))))).)))......(((.((.(.(((((((((.....)))))))))).)).))).. ( -28.00) >DroEre_CAF1 65932 116 - 1 AAUGAAGGAAAAUGAAGGGAAACCCAGCCAGACGGCCACCAU----CCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGCGCGCGAAAUU ..((((((........((....))..(((....)))......----...........))))))..(((.(((((.....)))))...(((((((((.....)))))))))....)))... ( -34.00) >DroYak_CAF1 67633 112 - 1 AAUGAAGGAAAACGAAGGAAAAUCCAGCCAGACGACCACCAU----CCAC----CCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGCGCGCGAAAUU ..((((((.....(((((....((......)).((......)----)...----...))))).))))))(((((.....)))))...(((((((((.....))))))))).......... ( -26.70) >consensus AGCGAAGGAAAAUGAAGGAAAAUCCAGCCAGACGACCACCAU____CCACCCACCCACCUUCACCUUCAGCCAGUGACUUUGGCACUGCACUUGAAAAUUUUUUAAGUGUGCGCGAAAUU ...(((((....((((((.......................................))))))))))).(((((.....)))))...(((((((((.....))))))))).......... (-26.07 = -26.03 + -0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:11 2006