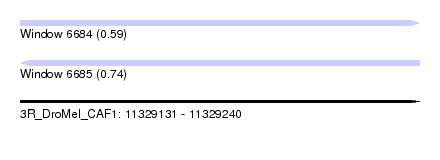
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,329,131 – 11,329,240 |
| Length | 109 |
| Max. P | 0.737680 |

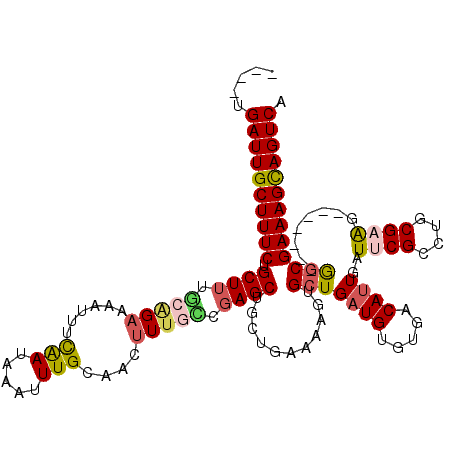
| Location | 11,329,131 – 11,329,240 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11329131 109 + 27905053 UGACUGCUUUCGCC-------UUUCGCAGGCGAAUCAAUGUCACACAUCAGCAAUUUUCAGAAGCUCGACAAAGUUGCAAAUUUAUUGAAAUUUUCUGCGCAGCAGAAAGCAAUCA---- .((.(((.((((((-------(.....)))))))((((((.....)....((((((((..(.....)...))))))))......)))))...(((((((...)))))))))).)).---- ( -27.80) >DroSec_CAF1 68284 109 + 1 UGACUGCUUUCGCU-------CCUCGCUGGCGAAUCAAUGUCACACAUCAGCACUUUUCAGCAGCUCGGAAAAGUUGCAAAUUUAUCGAAAUUUUCCGCAAAGCAGAAAGCAAUCA---- .((.((((((((((-------....(.(((((......))))))......((........((((((......))))))((((((....))))))...))..))).))))))).)).---- ( -25.60) >DroSim_CAF1 66480 109 + 1 UGACUGCUUUCGCU-------CCUCGCUGGCGAAUCAAUGUCACACAUCAGCACUUUUCAGCAGCUCGGAAAAGUUGCAAAUUUAUUGAAAUUUUCCGCAAAGCAGAAAGCAAUCA---- .((.(((((((...-------....(((((.((.....((........))....)).))))).((((((((((....(((.....)))...)))))))...))).))))))).)).---- ( -28.00) >DroEre_CAF1 63807 112 + 1 UGACUGCUUUCGUC-------CUUCGCAGGCGCAUCAAUGUCACACAUCAGCAAUUUUC-GCAGCUCGUCAAAGUUGCAACUUUAUUGAAAUUUUCGGAAAAGCAGAAAGCAAUCAGGUA (((.(((((((.((-------(...((....)).((((((.....)....((((((((.-((.....)).))))))))......))))).......)))......))))))).))).... ( -25.30) >DroYak_CAF1 65618 115 + 1 UGACUACUUUCGCCUUUUGCCUUUCGCAGGCGCAUCAGUGUCACACAUCAGCAGUUUUC-GCAGCUCGACAAAGUUGCAAAUUUAUUAAAAUUCGCUGAGAAGCAGAAAGCAAUCA---- ...........((.((((((..(((...(((((....))))).....(((((.......-((((((......)))))).(((((....))))).)))))))))))))).)).....---- ( -29.70) >consensus UGACUGCUUUCGCC_______CUUCGCAGGCGAAUCAAUGUCACACAUCAGCAAUUUUCAGCAGCUCGACAAAGUUGCAAAUUUAUUGAAAUUUUCCGCAAAGCAGAAAGCAAUCA____ (((.(((((((.................((((......))))........((........((((((......))))))((((((....))))))........)).))))))).))).... (-19.42 = -20.22 + 0.80)

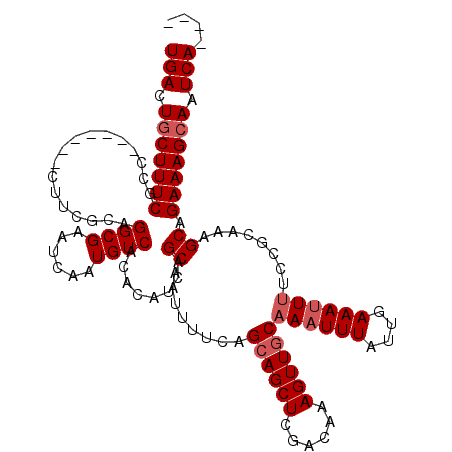
| Location | 11,329,131 – 11,329,240 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.70 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11329131 109 - 27905053 ----UGAUUGCUUUCUGCUGCGCAGAAAAUUUCAAUAAAUUUGCAACUUUGUCGAGCUUCUGAAAAUUGCUGAUGUGUGACAUUGAUUCGCCUGCGAAA-------GGCGAAAGCAGUCA ----.((((((((((.(((.(((((.((((((....))))))(((((..(((((.((.((...........)).)).)))))..).)).)))))))...-------))))))))))))). ( -34.50) >DroSec_CAF1 68284 109 - 1 ----UGAUUGCUUUCUGCUUUGCGGAAAAUUUCGAUAAAUUUGCAACUUUUCCGAGCUGCUGAAAAGUGCUGAUGUGUGACAUUGAUUCGCCAGCGAGG-------AGCGAAAGCAGUCA ----.((((((((((.(((((.(((((((.(((((.....))).)).))))))).((((.((((.(((((........).))))..)))).))))..))-------))))))))))))). ( -38.50) >DroSim_CAF1 66480 109 - 1 ----UGAUUGCUUUCUGCUUUGCGGAAAAUUUCAAUAAAUUUGCAACUUUUCCGAGCUGCUGAAAAGUGCUGAUGUGUGACAUUGAUUCGCCAGCGAGG-------AGCGAAAGCAGUCA ----.((((((((((.(((((.(((((((.(((((.....))).)).))))))).((((.((((.(((((........).))))..)))).))))..))-------))))))))))))). ( -38.80) >DroEre_CAF1 63807 112 - 1 UACCUGAUUGCUUUCUGCUUUUCCGAAAAUUUCAAUAAAGUUGCAACUUUGACGAGCUGC-GAAAAUUGCUGAUGUGUGACAUUGAUGCGCCUGCGAAG-------GACGAAAGCAGUCA .....((((((((((..(((((.((......(((((....(((((.(((....))).)))-))...(..(......)..).)))))......)).))))-------)..)))))))))). ( -27.90) >DroYak_CAF1 65618 115 - 1 ----UGAUUGCUUUCUGCUUCUCAGCGAAUUUUAAUAAAUUUGCAACUUUGUCGAGCUGC-GAAAACUGCUGAUGUGUGACACUGAUGCGCCUGCGAAAGGCAAAAGGCGAAAGUAGUCA ----.((((((((((.((((.(((((((((((....)))))))).....(((((.((.((-(.....)))....)).))))).)))...((((.....))))...)))))))))))))). ( -36.90) >consensus ____UGAUUGCUUUCUGCUUUGCAGAAAAUUUCAAUAAAUUUGCAACUUUGCCGAGCUGCUGAAAAGUGCUGAUGUGUGACAUUGAUUCGCCUGCGAAG_______GGCGAAAGCAGUCA .....((((((((((.((((.(((((......(((.....)))....))))).))))...........(((((((.....))))..((((....))))........))))))))))))). (-21.30 = -21.86 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:20:04 2006