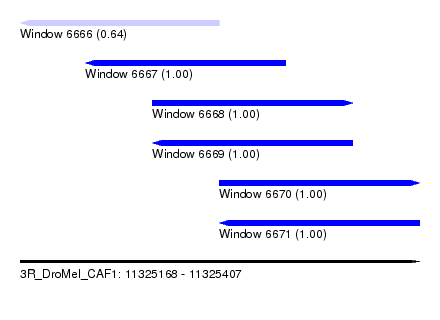
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,325,168 – 11,325,407 |
| Length | 239 |
| Max. P | 0.999895 |

| Location | 11,325,168 – 11,325,287 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.49 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.32 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11325168 119 - 27905053 UUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCCU-UUCCGCUCAACUUUUCGCAUUCGGCGCUUGGCAAAUUA ..((((...((((....))))..))))............((((((.(.((((((...........(((...(((...))).-..)))..........))))))).))))))......... ( -28.30) >DroSec_CAF1 64377 119 - 1 UUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCCU-UUCCGCUCAACUUUUCGCAUUCCGCGUUUGGCAAAUUA ..((((...((((....))))..))))............((((((((.((((((...........(((...(((...))).-..)))..........))))))))))))))......... ( -30.20) >DroSim_CAF1 60660 119 - 1 UUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCCU-UUCCGCUCAACUUUUCGCAUUCCGCGUUUGGCAAAUUA ..((((...((((....))))..))))............((((((((.((((((...........(((...(((...))).-..)))..........))))))))))))))......... ( -30.20) >DroEre_CAF1 59720 120 - 1 UUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCCUCUUCCGCUCAACUUUUCGCAUUCCGCGCUUGGCAAAUUA ..((((...((((....))))..))))............((((((((.((((((...........(((...(((...)))....)))..........))))))))))))))......... ( -31.90) >DroYak_CAF1 61447 119 - 1 UUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUAUGCAAUUUUUGGUCUGGGCUUCCCU-UUCCGCUCAACUUUUCGCAUUCCGCGUUUGGCAAAUUA ..((((...((((....))))..))))............((((((((.((((((...........(((...(((...))).-..)))..........))))))))))))))......... ( -30.20) >consensus UUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCCU_UUCCGCUCAACUUUUCGCAUUCCGCGUUUGGCAAAUUA ..((((...((((....))))..))))............((((((((.((((((...........(((...(((...)))....)))..........))))))))))))))......... (-29.36 = -29.32 + -0.04)

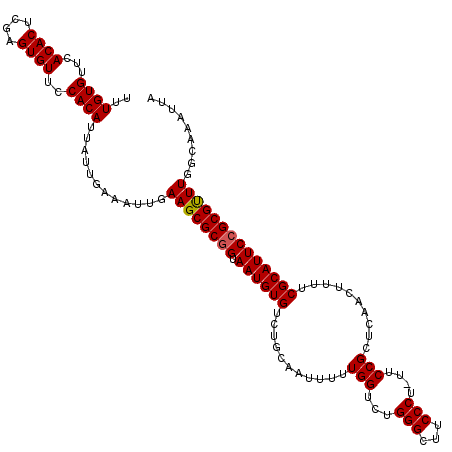

| Location | 11,325,207 – 11,325,327 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -37.20 |
| Energy contribution | -37.04 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11325207 120 - 27905053 UUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCC ...............(((((((((((((....(((((((....(((....)))....)))))))..))))))(((((((.((((((....)))).)).)))))))...)))))))..... ( -37.50) >DroSec_CAF1 64416 120 - 1 UUGGCCAACUAUUUCGCCUAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCC ..(((((((((((((.....)))))))))...(((((((....(((....)))....))))))).((.....(((((((.((((((....)))).)).))))))).....)))))).... ( -37.90) >DroSim_CAF1 60699 120 - 1 UUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCC ...............(((((((((((((....(((((((....(((....)))....)))))))..))))))(((((((.((((((....)))).)).)))))))...)))))))..... ( -37.50) >DroEre_CAF1 59760 120 - 1 UUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCC ...............(((((((((((((....(((((((....(((....)))....)))))))..))))))(((((((.((((((....)))).)).)))))))...)))))))..... ( -37.50) >DroYak_CAF1 61486 120 - 1 UUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUAUGCAAUUUUUGGUCUGGGCUUCCC ...............(((((((((((((....(((((((....(((....)))....)))))))..))))))(((((((..(((((....)))))...)))))))...)))))))..... ( -37.50) >consensus UUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGAAGCGCGGUAAUGUGUCUGCAAUUUUUGGUCUGGGCUUCCC ...............(((((((((((((....(((((((....(((....)))....)))))))..))))))(((((((..(((((....)))))...)))))))...)))))))..... (-37.20 = -37.04 + -0.16)



| Location | 11,325,247 – 11,325,367 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -33.84 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11325247 120 + 27905053 UCAAUUUCAAUAAUGUGGAACACUCGAGUGUGAACACAAACGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAU ...(((((.....((((..((((....))))...)))).....((((.((((....)))).)))))))))....((((((((....)))))))).....(((.........)))...... ( -33.90) >DroSec_CAF1 64456 120 + 1 UCAAUUUCAAUAAUGUGGAACACUCGAGUGUGAACACAAACGUGCUCCGAAAACUAUUUCUAGGCGAAAUAGUUGGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAU ....((((.....((((..((((....))))...))))....((((..((.(((((((((.....))))))))).(((((((....))))))).))...))))...)))).......... ( -32.80) >DroSim_CAF1 60739 120 + 1 UCAAUUUCAAUAAUGUGGAACACUCGAGUGUGAACACAAACGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAU ...(((((.....((((..((((....))))...)))).....((((.((((....)))).)))))))))....((((((((....)))))))).....(((.........)))...... ( -33.90) >DroEre_CAF1 59800 119 + 1 UCAAUUUCAAUAAUGUGGAACACUCGAGUGUGAACACAAACGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAG-AGAAAGGCCAAAAAU ....((((.....((((..((((....))))...))))......(((((.((((((((((.....))))))))))(((((((....))))))).....))).))-.)))).......... ( -34.70) >DroYak_CAF1 61526 120 + 1 UCAAUUUCAAUAAUGUGGAACACUCGAGUGUGAACACAAACGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAU ...(((((.....((((..((((....))))...)))).....((((.((((....)))).)))))))))....((((((((....)))))))).....(((.........)))...... ( -33.90) >consensus UCAAUUUCAAUAAUGUGGAACACUCGAGUGUGAACACAAACGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAU ....((((.....((((..((((....))))...))))....((((..((((((((((((.....))))))))))(((((((....))))))).))...))))...)))).......... (-33.12 = -33.32 + 0.20)


| Location | 11,325,247 – 11,325,367 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -41.56 |
| Energy contribution | -41.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.33 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11325247 120 - 27905053 AUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGA (((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..(((((((....(((....)))....)))))))................ ( -42.20) >DroSec_CAF1 64456 120 - 1 AUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCCAACUAUUUCGCCUAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGA (((((((((.........)))))))))(((((((....))))))).(((((((((.....)))))))))...(((((((....(((....)))....)))))))................ ( -40.40) >DroSim_CAF1 60739 120 - 1 AUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGA (((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..(((((((....(((....)))....)))))))................ ( -42.20) >DroEre_CAF1 59800 119 - 1 AUUUUUGGCCUUUCU-CUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGA (((((((((......-..)))))))))(((((((....)))))))((((((((((.....))))))))))..(((((((....(((....)))....)))))))................ ( -43.00) >DroYak_CAF1 61526 120 - 1 AUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGA (((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..(((((((....(((....)))....)))))))................ ( -42.20) >consensus AUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACGUUUGUGUUCACACUCGAGUGUUCCACAUUAUUGAAAUUGA (((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..(((((((....(((....)))....)))))))................ (-41.56 = -41.76 + 0.20)

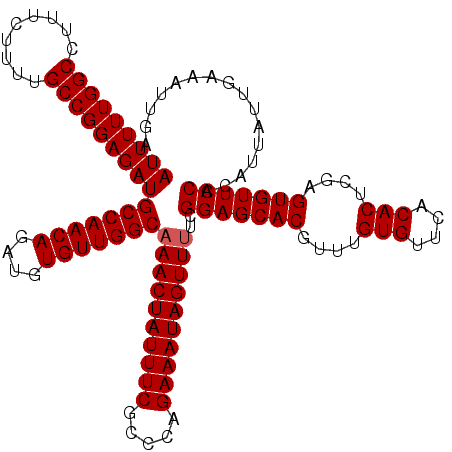
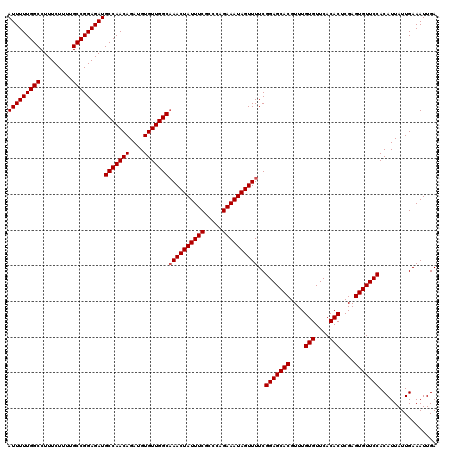
| Location | 11,325,287 – 11,325,407 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11325287 120 + 27905053 CGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAUUGGUAUUAUUGAACAGCAAUAAAUUUGACUAGGAACACAG .(((.(((((((((((((((.....))))))))))(((((((....))))))).))...((((((......((((.....)))).((((((.....)))))).)))).)).))).))).. ( -35.30) >DroSec_CAF1 64496 120 + 1 CGUGCUCCGAAAACUAUUUCUAGGCGAAAUAGUUGGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAUUGGUAUUAUUGAACAGCAAUAAAUUUGACUAGGAACACAG .(((.(((((.(((((((((.....))))))))).(((((((....))))))).))...((((((......((((.....)))).((((((.....)))))).)))).)).))).))).. ( -34.40) >DroSim_CAF1 60779 120 + 1 CGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAUUGGUAUUAUUGAACAGCAAUAAAUUUGACUAGGAACACAG .(((.(((((((((((((((.....))))))))))(((((((....))))))).))...((((((......((((.....)))).((((((.....)))))).)))).)).))).))).. ( -35.30) >DroEre_CAF1 59840 119 + 1 CGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAG-AGAAAGGCCAAAAAUUGGUAUUAUUGAACAGCAAUAAAUUUGACUAGGAACACAG .(((.(((((((((((((((.....))))))))))(((((((....))))))).))...(((((-(.....((((.....)))).((((((.....)))))).)))).)).))).))).. ( -35.40) >DroYak_CAF1 61566 120 + 1 CGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAUUGGUAUUAUUGAACAGCAAUAAAUUUGACUAGGAACACGG ((((.(((((((((((((((.....))))))))))(((((((....))))))).))...((((((......((((.....)))).((((((.....)))))).)))).)).))).)))). ( -37.00) >consensus CGUGCUCCGAAAACUAUUUCUGGGCGAAAUAGUUUGCCAACACAUCUGUUGGCAUCUCCGGCAAAAGAAAGGCCAAAAAUUGGUAUUAUUGAACAGCAAUAAAUUUGACUAGGAACACAG .(((.(((((((((((((((.....))))))))))(((((((....))))))).))...((((((......((((.....)))).((((((.....)))))).)))).)).))).))).. (-34.22 = -34.46 + 0.24)

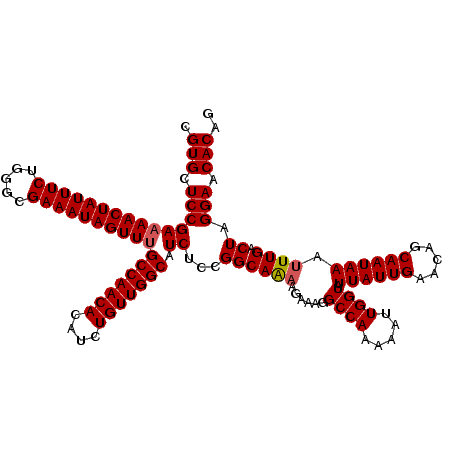
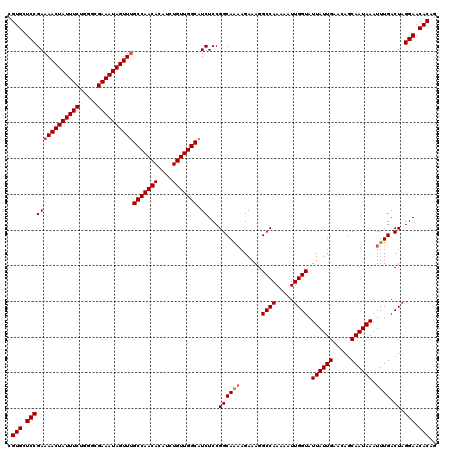
| Location | 11,325,287 – 11,325,407 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.76 |
| Consensus MFE | -39.12 |
| Energy contribution | -39.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.22 |
| SVM RNA-class probability | 0.999840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11325287 120 - 27905053 CUGUGUUCCUAGUCAAAUUUAUUGCUGUUCAAUAAUACCAAUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACG ..(((((((.........((((((.....)))))).....(((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..))))))). ( -39.70) >DroSec_CAF1 64496 120 - 1 CUGUGUUCCUAGUCAAAUUUAUUGCUGUUCAAUAAUACCAAUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCCAACUAUUUCGCCUAGAAAUAGUUUUCGGAGCACG ..(((((((.........((((((.....)))))).....(((((((((.........)))))))))(((((((....))))))).(((((((((.....)))))))))...))))))). ( -37.90) >DroSim_CAF1 60779 120 - 1 CUGUGUUCCUAGUCAAAUUUAUUGCUGUUCAAUAAUACCAAUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACG ..(((((((.........((((((.....)))))).....(((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..))))))). ( -39.70) >DroEre_CAF1 59840 119 - 1 CUGUGUUCCUAGUCAAAUUUAUUGCUGUUCAAUAAUACCAAUUUUUGGCCUUUCU-CUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACG ..(((((((..((((((...((((.((((....)))).)))).))))))...(((-(.....))))((((((((....))))))))(((((((((.....)))))))))...))))))). ( -40.80) >DroYak_CAF1 61566 120 - 1 CCGUGUUCCUAGUCAAAUUUAUUGCUGUUCAAUAAUACCAAUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACG .((((((((.........((((((.....)))))).....(((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..)))))))) ( -40.70) >consensus CUGUGUUCCUAGUCAAAUUUAUUGCUGUUCAAUAAUACCAAUUUUUGGCCUUUCUUUUGCCGGAGAUGCCAACAGAUGUGUUGGCAAACUAUUUCGCCCAGAAAUAGUUUUCGGAGCACG ..(((((((.........((((((.....)))))).....(((((((((.........)))))))))(((((((....)))))))((((((((((.....))))))))))..))))))). (-39.12 = -39.32 + 0.20)


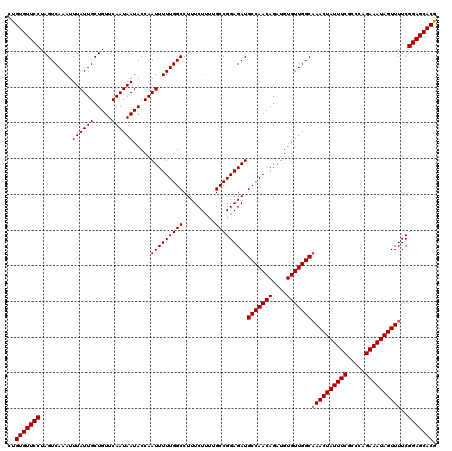
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:50 2006