
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,322,900 – 11,323,035 |
| Length | 135 |
| Max. P | 0.602179 |

| Location | 11,322,900 – 11,323,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -26.66 |
| Energy contribution | -26.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11322900 120 + 27905053 CAGGAUAAUUAAGAAAUUUUUCAGUUCCCGUCUCUGGACCUCACUUUUGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUU ............(((.((..((((((.((((....(((((((((....))).........))))))((((((((((.......))))))))))....)))).))))))..)).))).... ( -31.30) >DroSec_CAF1 62107 120 + 1 CAGGAUAAUUAAGAAAUUUUUCAGUUCCCGCCUCUGGACCCCACUUUAGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUU ..(((...((((..((((((.((.((((((((...(((((((((....))).........))))))...))))).(((.....)))....))).....))))))))..))))..)))... ( -32.50) >DroSim_CAF1 58389 120 + 1 CAGGAUAAUUAAGAAAUUUUUCAGUUCCCGCCUCUGGACCCCACUUUUGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUU ..(((...((((..((((((.((.((((((((...(((((((((....))).........))))))...))))).(((.....)))....))).....))))))))..))))..)))... ( -32.80) >DroEre_CAF1 57415 120 + 1 CAGGAUAAUUAAGAAAUUUUUCAGUUCCCGCCUCUGGACCCCACUUUCCUGGCCCUCGUUGGGUCCUUUGGUGGUGCGACUUUCGCUAUCGAAGUAAAUGGAAAUUGAUUGAAUUCCAUU ..(((...((((..(((((((.(.((((((((...((((((.((.............)).))))))...))))).(((.....)))....))).)....)))))))..))))..)))... ( -32.92) >DroYak_CAF1 59014 120 + 1 CAGGAUAAUUAAGAAAUUUUUCAGUUCCCGCCUCUGGACCCCACUUUCCUGGCCCUCAUUGGGAACUUUGGUGGUGCGACUUUGGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUU ..(((...((((..((((((.(((((((((((...(((........))).))).......))))))(((((((((.........))))))))).....))))))))..))))..)))... ( -29.21) >consensus CAGGAUAAUUAAGAAAUUUUUCAGUUCCCGCCUCUGGACCCCACUUUCGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUU ..(((...((((..((((((((((.........)))))..(((.......(((((.....))))).((((((((((.......)))))))))).....))))))))..))))..)))... (-26.66 = -26.86 + 0.20)

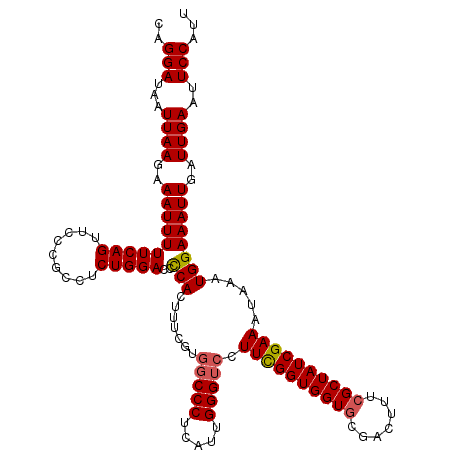

| Location | 11,322,940 – 11,323,035 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.39 |
| Mean single sequence MFE | -34.51 |
| Consensus MFE | -27.86 |
| Energy contribution | -27.78 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11322940 95 + 27905053 UCACUUUUGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUUAGGCCAGGAAGGGAA------------------------- ((.(((((.(((((............((((((((((.......))))))))))...(((((((.((....)).))))))).))))).))))))).------------------------- ( -32.50) >DroSec_CAF1 62147 120 + 1 CCACUUUAGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUUAGGCCAGGAAGGGAAGCAGGCGAAAUCGAACCAAGCCCAA ((((....))))((((..((((..((((((((((((.......))))))))))...(((((((.((....)).))))))).))))))..)))).....(((.............)))... ( -35.92) >DroSim_CAF1 58429 120 + 1 CCACUUUUGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUUAGGCCAGGAAGGAAAGCAGGCGAAAUCGAACCAAGCCAAA ...(((((.(((((............((((((((((.......))))))))))...(((((((.((....)).))))))).))))).)))))......(((.............)))... ( -35.92) >DroEre_CAF1 57455 115 + 1 CCACUUUCCUGGCCCUCGUUGGGUCCUUUGGUGGUGCGACUUUCGCUAUCGAAGUAAAUGGAAAUUGAUUGAAUUCCAUUAGGCCAGCAUAGGCAGCAGGCGAAAUCGAACCGC-----A ..........(((((.....))))).....(((((.(((.(((((((.........(((((((.((....)).)))))))..(((......)))....)))))))))).)))))-----. ( -38.30) >DroYak_CAF1 59054 115 + 1 CCACUUUCCUGGCCCUCAUUGGGAACUUUGGUGGUGCGACUUUGGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUUAGGCCAGCAAAGGAAGCAGGCGAAAUCGAACCAA-----A (((((.......(((.....)))......))))).......((((...((((....(((((((.((....)).)))))))..(((.((.......)).)))....)))).))))-----. ( -29.92) >consensus CCACUUUCGUGGCCCUCAUUGGGUCCUUCGGUGGUGCGACUUUCGCUAUCGAAAUAAAUGGAAAUUGAUUGAAUUCCAUUAGGCCAGGAAGGGAAGCAGGCGAAAUCGAACCAA_____A ...((((..(((((............((((((((((.......))))))))))...(((((((.((....)).))))))).)))))..))))............................ (-27.86 = -27.78 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:38 2006