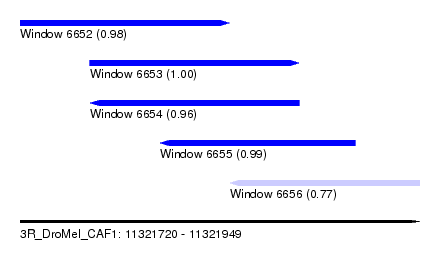
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,321,720 – 11,321,949 |
| Length | 229 |
| Max. P | 0.997994 |

| Location | 11,321,720 – 11,321,840 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -20.78 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321720 120 + 27905053 GGAUUGGCGGUUCAAAUAAAUAAUGAAAACAAAGGGCGCAUAAAUAUUUGAAGAUCGCAAACCGAAUGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUU .((((.((((((((..(((((((((....(....)...)))...))))))....(((.....))).))))))....(.((((((....)))))).).......)).)))).......... ( -24.40) >DroSec_CAF1 60944 120 + 1 GGAUUGGCGGUUCAAAUAAAUAAUGAAAACAAAGGGCGCAUAAAUAUUUGAAGAUCGCAAACCGAGCGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUU .((((.((((((((((((....(((....(....)...)))...)))))))...((((.......)))))))....(.((((((....)))))).).......)).)))).......... ( -25.70) >DroSim_CAF1 57189 120 + 1 GGAUUGGCGGUUCAAAUAAAUAAUGAAAACAAAGGGCGCAUAAAUAUUUGAAGAUCGCAAACCGAGCGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUU .((((.((((((((((((....(((....(....)...)))...)))))))...((((.......)))))))....(.((((((....)))))).).......)).)))).......... ( -25.70) >DroEre_CAF1 56207 120 + 1 GGAUUCGCGAGUCAAAUAAAUAAUGAAAACAAAGGGCGCAUAAAUAUUUGAAGAUCGCAAGCCGAAUGAACCGAAACGGCUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUU .(((((((((.(((((((....(((....(....)...)))...)))))))...))))..................(.((((((....)))))).).........))))).......... ( -25.90) >DroYak_CAF1 57723 120 + 1 GGAUUCGCGGGUCAAAUAAAUAAUGAAAACAAAGGGCGCAUAAAUAUUUGAAGAUCGCAAAGCGAAUGAAUCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUU .(((((((((.(((((((....(((....(....)...)))...)))))))...))))....(((.....)))...(.((((((....)))))).).........))))).......... ( -25.10) >consensus GGAUUGGCGGUUCAAAUAAAUAAUGAAAACAAAGGGCGCAUAAAUAUUUGAAGAUCGCAAACCGAAUGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUU ((....((((.(((((((....(((....(....)...)))...)))))))...))))...)).............(.((((((....)))))).)....(((.....)))......... (-20.78 = -20.66 + -0.12)


| Location | 11,321,760 – 11,321,880 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -26.36 |
| Energy contribution | -25.56 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321760 120 + 27905053 UAAAUAUUUGAAGAUCGCAAACCGAAUGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGCAAA ......((((......((((((..............(.((((((....)))))).).........(((((((((....))).))))))))))))...(((((.......)))))..)))) ( -26.40) >DroSec_CAF1 60984 120 + 1 UAAAUAUUUGAAGAUCGCAAACCGAGCGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGCAAA ...............(((.....((((((((.....(.((((((....)))))).).........(((((((((....))).))))))))))))))..((((.......))))))).... ( -30.60) >DroSim_CAF1 57229 120 + 1 UAAAUAUUUGAAGAUCGCAAACCGAGCGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGCAAA ...............(((.....((((((((.....(.((((((....)))))).).........(((((((((....))).))))))))))))))..((((.......))))))).... ( -30.60) >DroEre_CAF1 56247 120 + 1 UAAAUAUUUGAAGAUCGCAAGCCGAAUGAACCGAAACGGCUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGCAAA ................((..(((...............((((((....))))))((((((((((((((((((((....))).)))))))))))..))))))...........)))))... ( -27.90) >DroYak_CAF1 57763 120 + 1 UAAAUAUUUGAAGAUCGCAAAGCGAAUGAAUCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAUUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGCAAA ...............(((((((((((((((..(((((.((((((....)))))).)....(((.....))).......))))...)))))))))))).((((.......))))))).... ( -32.00) >consensus UAAAUAUUUGAAGAUCGCAAACCGAAUGAACCGAAACGACUUACGAAAGUAGGUGGGAAACAAGUGAAUUGAAAAUUAUUUCCAAUUCGUUUGCUUUUCUCAUUAUAAUUGAGGCGCAAA ...............((((((.((((((((..(((((.((((((....)))))).)....(((.....))).......))))...)))))))).))).((((.......))))))).... (-26.36 = -25.56 + -0.80)


| Location | 11,321,760 – 11,321,880 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -23.96 |
| Consensus MFE | -22.74 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321760 120 - 27905053 UUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUUUCGGUUCAUUCGGUUUGCGAUCUUCAAAUAUUUA (((((..((((.......))))....))))).(((((((((.....)))))))))...................((((((..((............))..)))))).............. ( -23.00) >DroSec_CAF1 60984 120 - 1 UUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUUUCGGUUCGCUCGGUUUGCGAUCUUCAAAUAUUUA (((((..((((.......))))....))))).(((((((((.....)))))))))...................((((((..((...((...))..))..)))))).............. ( -23.30) >DroSim_CAF1 57229 120 - 1 UUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUUUCGGUUCGCUCGGUUUGCGAUCUUCAAAUAUUUA (((((..((((.......))))....))))).(((((((((.....)))))))))...................((((((..((...((...))..))..)))))).............. ( -23.30) >DroEre_CAF1 56247 120 - 1 UUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGCCGUUUCGGUUCAUUCGGCUUGCGAUCUUCAAAUAUUUA (((((..((((.......))))....))))).(((((((((.....)))))))))...................((((((((((............)))))))))).............. ( -29.70) >DroYak_CAF1 57763 120 - 1 UUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAAUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUUUCGAUUCAUUCGCUUUGCGAUCUUCAAAUAUUUA .((((..((((.......)))).(((((((((((...((((((((............))))))))....(((....)))..)))))).((.....))))))))))).............. ( -20.50) >consensus UUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAAGUCGUUUCGGUUCAUUCGGUUUGCGAUCUUCAAAUAUUUA (((((..((((.......))))....))))).(((((((((.....)))))))))...................((((((((((............)))))))))).............. (-22.74 = -22.86 + 0.12)



| Location | 11,321,800 – 11,321,912 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -23.32 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321800 112 - 27905053 AGGA------GAUACCGAUUCCGAGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA .(((------((((........(((.(((((((....))))))).)))......................(((((((((.....)))))))))...)))))))............... ( -29.20) >DroSec_CAF1 61024 112 - 1 AGGA------GAUACCGAUUCCGCGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA .(((------((((........((...((((((....))))))..((((.......))))....))....(((((((((.....)))))))))...)))))))............... ( -29.20) >DroSim_CAF1 57269 112 - 1 AGGA------GAUACCGAUUCCGCGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA .(((------((((........((...((((((....))))))..((((.......))))....))....(((((((((.....)))))))))...)))))))............... ( -29.20) >DroEre_CAF1 56287 118 - 1 UGCCGAUUCCGAUUCCGAUUCCCAGUUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA ...............(((.....(((((.(((.(((....))).)))))))).......(..((((((..(((((((((.....)))))))))..))))))..)........)))... ( -23.90) >DroYak_CAF1 57803 109 - 1 A---------GACACCGAUUCCGAGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAAUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA .---------.....(((....(((.(((((((....))))))).)))...........(..((((((..(((.(((((.....))))).)))..))))))..)........)))... ( -23.60) >consensus AGGA______GAUACCGAUUCCGAGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGAAAUAAUUUUCAAUUCACUUGUUUCCCACCUACUUUCGUAA ...............(((....(((.(((((((....))))))).)))...........(..((((((..(((((((((.....)))))))))..))))))..)........)))... (-23.32 = -24.12 + 0.80)



| Location | 11,321,840 – 11,321,949 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 88.11 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321840 109 - 27905053 CAGCAGUCGAAGGGGAUACCAGCUGGGUAACCGGAGG---AGGA------GAUACCGAUUCCGAGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGA ...((((((..((.....)).(((((....))((((.---.((.------....))..))))(((.(((((((....))))))).)))...............)))...)).)))).. ( -32.40) >DroSec_CAF1 61064 112 - 1 CAGCAGUCGACGGGGAUACCAGCUGGGGAACCGGAGGAGGAGGA------GAUACCGAUUCCGCGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGA ..((.((((.(((.....))..((((....)))).((((..((.------....))..)))))))))((((((....))))))..((((.......))))....))............ ( -33.30) >DroSim_CAF1 57309 112 - 1 CAGCAGUCGAAGGGGAUACCAGCUGGGGAACCGGAGGAGGAGGA------GAUACCGAUUCCGCGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGA ...((((((..((.....))..((((....)))).((((..((.------....))..))))((...((((((....))))))..((((.......))))....))...)).)))).. ( -33.20) >DroEre_CAF1 56327 115 - 1 C---AGUCGAAGGGGAAACAGGCCGAGGAACCGGAGGAGAUGCCGAUUCCGAUUCCGAUUCCCAGUUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGA (---(((((...(((((...((....((((.(((........))).))))....))..))))).(((((((((....))))))..((((.......))))...)))...)).)))).. ( -35.10) >DroYak_CAF1 57843 109 - 1 CAGCAGUCGAAGGGGAAACUGGCUGAGGAACCGGAGGAGGA---------GACACCGAUUCCGAGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAAUGGA ((....(((...((....)).(((..((((.(((....(..---------..).))).))))(((.(((((((....))))))).)))...............)))...)))..)).. ( -33.00) >consensus CAGCAGUCGAAGGGGAUACCAGCUGGGGAACCGGAGGAGGAGGA______GAUACCGAUUCCGAGAUGCAGGGGCAACUUUGCGCCUCAAUUAUAAUGAGAAAAGCAAACGAAUUGGA .....(((....((....))..((((....))))................))).(((((((......((((((....))))))((((((.......))))....))....))))))). (-26.96 = -27.32 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:35 2006