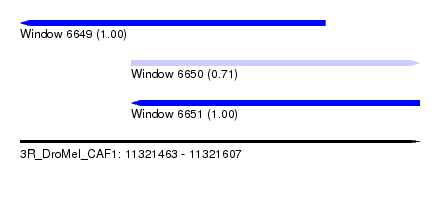
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,321,463 – 11,321,607 |
| Length | 144 |
| Max. P | 0.999792 |

| Location | 11,321,463 – 11,321,573 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.30 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321463 110 - 27905053 ----------AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAUUUAUGUAAGGCCAAAAUCAGAGGGCAGCCAAAUGCAUAAC ----------......(((((((((((((((((((((.......))))))))..))))))))(((......((((.....)))).....))).........)))))((.....))..... ( -35.20) >DroSec_CAF1 60704 105 - 1 ----------AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAUUUAUGUAAGGCCAAAAGCAGAG-----CCAAAUGCAUAAC ----------......((.((((((((((((((((((.......))))))))..))))))))))))..............(((((((.((((.......).)-----))...))))))). ( -35.70) >DroSim_CAF1 56944 110 - 1 ----------AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAUUUAUGUAAGGCCAAAAGCAGAGGGCGGGCAAAUGCAUAAC ----------......((.((((((((((((((((((.......))))))))..))))))))))))..............(((((((..(((....((.....)).)))...))))))). ( -40.20) >DroEre_CAF1 55929 120 - 1 GGAUCCGACGAUCCGACGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCAAGCUCCUGCGGCAAUAUAAUGAAUUAUUUAUGUAAGGCCAAAAACAGAGGGCAGCCAAAUGCAUAAC (((((....)))))...((((((((((((((((((((.......))))))))..))))))))(((.((((((........))))))...))).........)))).((.....))..... ( -43.10) >DroYak_CAF1 57459 110 - 1 ----------AUCUGCAGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUACAAUGAAUUAUUUAUGUAGGGCCAAAGACAGAGGGCAGCGAAAUGCAUAAC ----------...(((.((((((((((((((((((((.......))))))))..))))))))(((..((((.(((.....)))))))..))).........)))).)))........... ( -39.70) >consensus __________AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAUUUAUGUAAGGCCAAAAACAGAGGGCAGCCAAAUGCAUAAC .................((((((((((((((((((((.......))))))))..))))))))(((......((((.....)))).....))).........))))............... (-31.30 = -31.90 + 0.60)


| Location | 11,321,503 – 11,321,607 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -19.68 |
| Energy contribution | -19.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321503 104 + 27905053 AUAAUUCAUUAAAUUGCCGCAGGAGCUGGGCUUAUCGUUCGCAAACAAGCCACUCCUACGGACAUGAGAU----------------CCUCGGCUCCUAAAAAGCAUUAACAGCUUUAUCA ....................(((((((((((((((.(((((.................))))))))))..----------------.))))))))))..(((((.......))))).... ( -26.33) >DroSec_CAF1 60739 104 + 1 AUAAUUCAUUAAAUUGCCGCAGGAGCUGGGCUUAUCGUUCGCAAACAAGCCACUCCUACGGACAUGAGAU----------------CCUCGGCUCCUAAAAAACAUUAACAGCUUUAUCA ....................(((((((((((((((.(((((.................))))))))))..----------------.))))))))))....................... ( -22.93) >DroSim_CAF1 56984 104 + 1 AUAAUUCAUUAAAUUGCCGCAGGAGCUGGGCUUAUCGUUCGCAAACAAGCCACUCCUACGGACAUGAGAU----------------CCUCGGCUCCUAAAAAACAUUAACAGCUUUAUCA ....................(((((((((((((((.(((((.................))))))))))..----------------.))))))))))....................... ( -22.93) >DroEre_CAF1 55969 120 + 1 AUAAUUCAUUAUAUUGCCGCAGGAGCUUGGCUUAUCGUUCGCAAACAAGCCACUCCUACGGACGUCGGAUCGUCGGAUCCUCGGCUCCUCGGCUCCUAAAAAACAUUAACAGCUUUAUCA ...............((((.((((((.((((((...(((....)))))))))..........((..(((((....))))).))))))))))))........................... ( -32.50) >DroYak_CAF1 57499 104 + 1 AUAAUUCAUUGUAUUGCCGCAGGAGCUGGGCUUAUCGUUCGCAAACAAGCCACUCCUACGGACUGCAGAU----------------CCUCGGCUCCUAAAAAACAUUAACAGCUUUAUCA .........((((...(((.(((((...(((((...(((....)))))))).))))).)))..))))(((----------------....((((..(((......)))..))))..))). ( -23.90) >consensus AUAAUUCAUUAAAUUGCCGCAGGAGCUGGGCUUAUCGUUCGCAAACAAGCCACUCCUACGGACAUGAGAU________________CCUCGGCUCCUAAAAAACAUUAACAGCUUUAUCA ................(((.(((((...(((((...(((....)))))))).))))).))).............................((((..(((......)))..))))...... (-19.68 = -19.68 + -0.00)



| Location | 11,321,503 – 11,321,607 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.09 |
| SVM RNA-class probability | 0.999792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11321503 104 - 27905053 UGAUAAAGCUGUUAAUGCUUUUUAGGAGCCGAGG----------------AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAU ....(((((.......)))))......(((..((----------------((.....))))((((((((((((((((.......))))))))..)))))))))))............... ( -34.80) >DroSec_CAF1 60739 104 - 1 UGAUAAAGCUGUUAAUGUUUUUUAGGAGCCGAGG----------------AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAU .(((...(((.((((......)))).))).....----------------)))...((.((((((((((((((((((.......))))))))..)))))))))))).............. ( -34.40) >DroSim_CAF1 56984 104 - 1 UGAUAAAGCUGUUAAUGUUUUUUAGGAGCCGAGG----------------AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAU .(((...(((.((((......)))).))).....----------------)))...((.((((((((((((((((((.......))))))))..)))))))))))).............. ( -34.40) >DroEre_CAF1 55969 120 - 1 UGAUAAAGCUGUUAAUGUUUUUUAGGAGCCGAGGAGCCGAGGAUCCGACGAUCCGACGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCAAGCUCCUGCGGCAAUAUAAUGAAUUAU .......(((.((((......)))).)))......(((..(((((....))))).......((((((((((((((((.......))))))))..)))))))))))............... ( -40.00) >DroYak_CAF1 57499 104 - 1 UGAUAAAGCUGUUAAUGUUUUUUAGGAGCCGAGG----------------AUCUGCAGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUACAAUGAAUUAU .(((...(((.((((......)))).))).....----------------)))....(.((((((((((((((((((.......))))))))..)))))))))))............... ( -33.60) >consensus UGAUAAAGCUGUUAAUGUUUUUUAGGAGCCGAGG________________AUCUCAUGUCCGUAGGAGUGGCUUGUUUGCGAACGAUAAGCCCAGCUCCUGCGGCAAUUUAAUGAAUUAU .......(((.((((......)))).)))............................(.((((((((((((((((((.......))))))))..)))))))))))............... (-32.70 = -32.70 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:30 2006