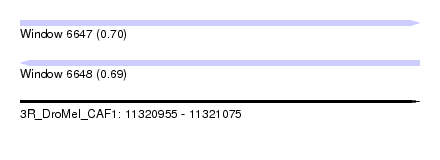
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,320,955 – 11,321,075 |
| Length | 120 |
| Max. P | 0.699331 |

| Location | 11,320,955 – 11,321,075 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -47.98 |
| Consensus MFE | -33.93 |
| Energy contribution | -35.37 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11320955 120 + 27905053 GAACUCUUCGCCACCUACGGCAACAUCGUGGAUGUGAACUUGCUGCGCCACAAGUUCAACAAUAGGUCCCGUGGCGUGGCCUUCUUGCAAUUUGAGUUGGUCCGCGACGCCGAGGUGGCC .........(((((((.((((....((((((((..(((...((..((((((....((.......))....))))))..)).)))...((((....)))))))))))).))))))))))). ( -51.00) >DroSec_CAF1 60178 120 + 1 GAGCUCUUUGCCCCCUUCGGCAACAUCUUGGAUGUGACCUUGCUGCGCCACAGGUUCAACAAUAAGUUCCGUGGCGUGGCCUUCUUGGAUUUCGAGCUGGUCCGCGACGCCGAGGAGGCC .........(((.(((.((((.(((((...)))))((((..((..((((((.((..(........)..))))))))..))...((((.....))))..))))......))))))).))). ( -46.80) >DroSim_CAF1 56418 120 + 1 GAGCUCUUUGCCCCCUACGGCAAACUCUUGGAUGUGACCUUGCUGCGCCACAGGUUCAACAAUAAGUUCCGUGGCGUGGCCUUCUUGGAUUUCGAGCUGGCCCGCGACGCCGAGGAGGCC .........(((.(((.((((........((......))((((.(((((((.((..(........)..)))))))))((((..((((.....))))..)))).)))).))))))).))). ( -46.50) >DroEre_CAF1 55391 120 + 1 GAGCUCUUCGCCGUCUACGGCAUCGUCUUGGAUGUGAACUUGCUGCGCCACAAGUUCAACCGAACUUCCCGCGGAGUGGCCUUCGUGCACUUCAAGCACUGGCGGGACGCCGAGAUGGCC .........(((((((.((((...((.((((...((((((((........)))))))).)))))).((((((((((....))))((((.......))))..)))))).))))))))))). ( -52.10) >DroYak_CAF1 56877 120 + 1 GAGCUCUUCGCCACCUACGGCAACGUCUUGGAUAUCAAUUUGCUGCGCCACAAGUUCAGCAAAAUGUCCCGUGGCGUGGCCUUCGUACAUUUCGAGCACUGCCGGGACGCCGAGAUGGCC ..((.....(((......)))..((((((((.......(((((((.((.....)).)))))))..(((((..((((..((..(((.......)))))..))))))))).)))))))))). ( -43.50) >consensus GAGCUCUUCGCCACCUACGGCAACAUCUUGGAUGUGAACUUGCUGCGCCACAAGUUCAACAAUAAGUCCCGUGGCGUGGCCUUCUUGCAUUUCGAGCUGGGCCGCGACGCCGAGGUGGCC .........(((((((.((((....((((((..........((..((((((...................))))))..))...((((.....)))).....)))))).))))))))))). (-33.93 = -35.37 + 1.44)



| Location | 11,320,955 – 11,321,075 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.50 |
| Mean single sequence MFE | -47.82 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.28 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11320955 120 - 27905053 GGCCACCUCGGCGUCGCGGACCAACUCAAAUUGCAAGAAGGCCACGCCACGGGACCUAUUGUUGAACUUGUGGCGCAGCAAGUUCACAUCCACGAUGUUGCCGUAGGUGGCGAAGAGUUC .(((((((((((((((.(((............((((..(((((.((...)))).))).))))(((((((((......)))))))))..))).))))...)))).)))))))......... ( -48.20) >DroSec_CAF1 60178 120 - 1 GGCCUCCUCGGCGUCGCGGACCAGCUCGAAAUCCAAGAAGGCCACGCCACGGAACUUAUUGUUGAACCUGUGGCGCAGCAAGGUCACAUCCAAGAUGUUGCCGAAGGGGGCAAAGAGCUC .((((((((((((.....((((...((.........))..((..(((((((((((.....)))....))))))))..))..))))(((((...)))))))))).)))))))......... ( -46.10) >DroSim_CAF1 56418 120 - 1 GGCCUCCUCGGCGUCGCGGGCCAGCUCGAAAUCCAAGAAGGCCACGCCACGGAACUUAUUGUUGAACCUGUGGCGCAGCAAGGUCACAUCCAAGAGUUUGCCGUAGGGGGCAAAGAGCUC .(((((((((((((.(((((((..((.(.....).))..)))).))).)).((((((.((((((..((...))..))))))((......))..)))))))))).)))))))......... ( -49.20) >DroEre_CAF1 55391 120 - 1 GGCCAUCUCGGCGUCCCGCCAGUGCUUGAAGUGCACGAAGGCCACUCCGCGGGAAGUUCGGUUGAACUUGUGGCGCAGCAAGUUCACAUCCAAGACGAUGCCGUAGACGGCGAAGAGCUC .(((.((((((((((((((..((((.......))))...((.....)))))))).(((.((.(((((((((......)))))))))...))..)))..))))).))).)))......... ( -47.80) >DroYak_CAF1 56877 120 - 1 GGCCAUCUCGGCGUCCCGGCAGUGCUCGAAAUGUACGAAGGCCACGCCACGGGACAUUUUGCUGAACUUGUGGCGCAGCAAAUUGAUAUCCAAGACGUUGCCGUAGGUGGCGAAGAGCUC .(((((((((((((((((...((((.......))))...(((...))).))))))..(((((((..(.....)..))))))).................)))).)))))))......... ( -47.80) >consensus GGCCACCUCGGCGUCGCGGACCAGCUCGAAAUGCAAGAAGGCCACGCCACGGGACUUAUUGUUGAACUUGUGGCGCAGCAAGUUCACAUCCAAGACGUUGCCGUAGGGGGCGAAGAGCUC .((((((((((((((..(((.....((.........))..((..(((((((((.............)))))))))..)).........)))..)))...)))).)))))))......... (-34.20 = -34.28 + 0.08)

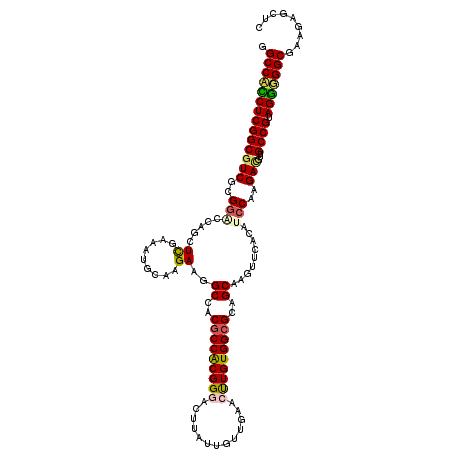

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:27 2006