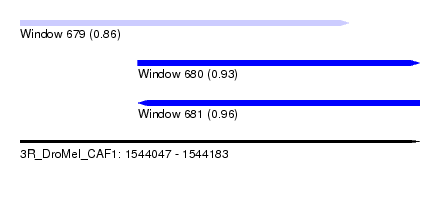
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,544,047 – 1,544,183 |
| Length | 136 |
| Max. P | 0.955594 |

| Location | 1,544,047 – 1,544,159 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -26.51 |
| Energy contribution | -27.25 |
| Covariance contribution | 0.74 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
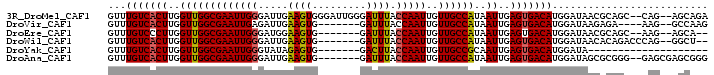
>3R_DroMel_CAF1 1544047 112 + 27905053 GCAGAGCAGAGUACAGGCUGCAGGAUCAGGCCUGGAUAUGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGAUUUACCAAUUGUUGCCAUAAUUGAGUGA (((((.((.....((((((.........))))))....)).)))))((((..(((((((((((........)))..((((((......)))))).))))))..))..)))). ( -35.30) >DroSec_CAF1 45595 112 + 1 GUAGAGCAGAGUACAGGCUGCAGGAUCAGGCCAGGAUAUGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGAUUUACCAAUUGUUGCCAUAAUUGAGUGA .....((((........)))).....(((((((.....)))))))(((((..(((((((((((........)))..((((((......)))))).))))))..))..))))) ( -33.50) >DroSim_CAF1 46234 112 + 1 GUAGAGCAGAGUACAGGCUGCAGGAUCAGGCCAGGAUAUGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGACUUACCAAUUGUUGCCAUAAUUGAGUGA .....((((........)))).....(((((((.....)))))))(((((..(((((((((((((.....((((.........)))).)))))..))))))..))..))))) ( -33.60) >DroEre_CAF1 44978 103 + 1 --AGCGCAGGGAUCGGGCGGCAGGAUCAGGCCAGGACAUGGUUUGUCCCUUGGUUGGCGAAUUGGGAUGGAAGUG-------GAUUUACCAAUUGUUGCCAUAAUUGAGUGA --.((.(((......((((((((((.(((((((.....))))))))))....(((((..((((.(........).-------))))..))))))))))))....))).)).. ( -31.60) >DroYak_CAF1 40988 103 + 1 --AGAGCAGAGAACAGGCUGCAGGAUCAGACCAGGACAUGGUUUGUCACUUGGUUGGCGAAUUGGGUAUAGAGUG-------GACUUACCAAUUGUUGCCGCAAUUGAGUGA --...((((........)))).....(((((((.....)))))))(((((..((((((((....((((.((....-------..)))))).....)))))..)))..))))) ( -33.10) >DroAna_CAF1 43455 87 + 1 ------------------GUCCUGAGGCGGCCAGGACUUGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUG-------GAUUUACCAAUUGUUGCCAUAAUUGAGUGA ------------------((((((.......))))))........(((((..(((((((((((((..........-------......)))))..))))))..))..))))) ( -28.39) >consensus __AGAGCAGAGUACAGGCUGCAGGAUCAGGCCAGGACAUGGUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUG_______GAUUUACCAAUUGUUGCCAUAAUUGAGUGA .....((((........)))).....(((((((.....)))))))(((((..(((((((((((((.....((((.........)))).)))))..))))))..))..))))) (-26.51 = -27.25 + 0.74)

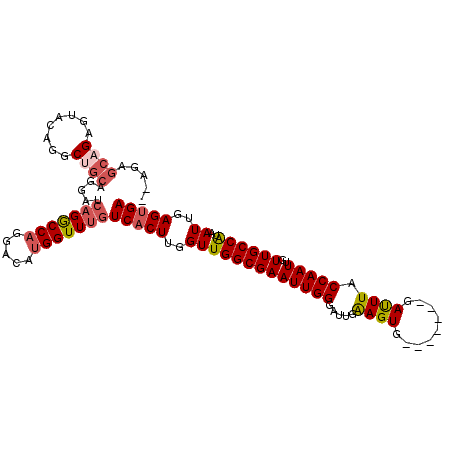
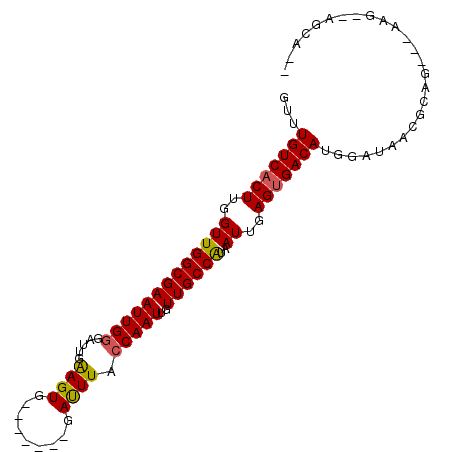
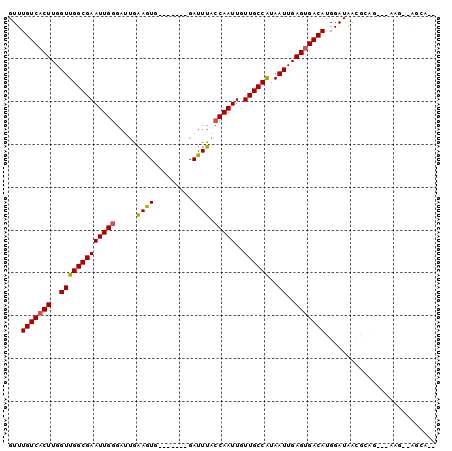
| Location | 1,544,087 – 1,544,183 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -25.16 |
| Consensus MFE | -22.51 |
| Energy contribution | -22.43 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1544087 96 + 27905053 GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUGGGAUUGGGAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGC--CAG--AGCAGA ((((((((((..(((((((((((........)))..((((((......)))))).))))))..))..)))))).(((.........)--)))--)))... ( -28.10) >DroVir_CAF1 55436 87 + 1 GUUUGUCACUUGGUUGGCGAAUUGAGAUUGAAGUG-------GAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAAGAGA----AAG--GCCAAG ...(((((((..((((((((.....(((((.....-------.......))))).))))))..))..)))))))(((........----...--.))).. ( -23.00) >DroEre_CAF1 45016 87 + 1 GUUUGUCCCUUGGUUGGCGAAUUGGGAUGGAAGUG-------GAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAGC--AAG--AGCA-- ...((((.((..(((((((((((((....(((...-------..))).)))))..))))))..))..)).))))...........((--...--.)).-- ( -21.30) >DroWil_CAF1 46386 89 + 1 GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUG-------GAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACACAGACCCAG--GGCU-- ...(((((((..(((((((((((((..........-------......)))))..))))))..))..)))))))(((...........))).--....-- ( -25.29) >DroYak_CAF1 41026 73 + 1 GUUUGUCACUUGGUUGGCGAAUUGGGUAUAGAGUG-------GACUUACCAAUUGUUGCCGCAAUUGAGUGACAUGGAUA-------------------- ...(((((((..((((((((....((((.((....-------..)))))).....)))))..)))..)))))))......-------------------- ( -25.00) >DroAna_CAF1 43477 91 + 1 GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUG-------GAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAGCGCGGG--GAGCGAGCGGG ((((((((((..(((((((((((((..........-------......)))))..))))))..))..))))))).......(((...--..))))))... ( -28.29) >consensus GUUUGUCACUUGGUUGGCGAAUUGGGAUUGAAGUG_______GAUUUACCAAUUGUUGCCAUAAUUGAGUGACAUGGAUAACGCAG___AAG__AGCA__ ...(((((((..(((((((((((((.....((((.........)))).)))))..))))))..))..))))))).......................... (-22.51 = -22.43 + -0.08)
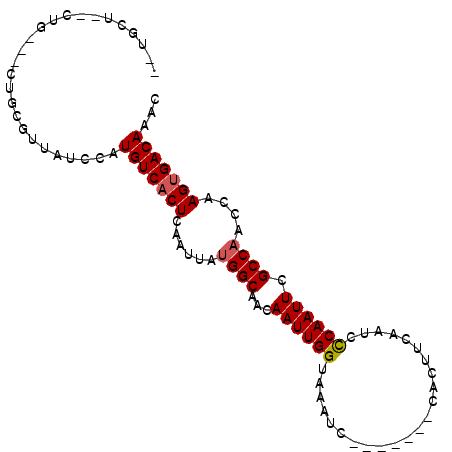
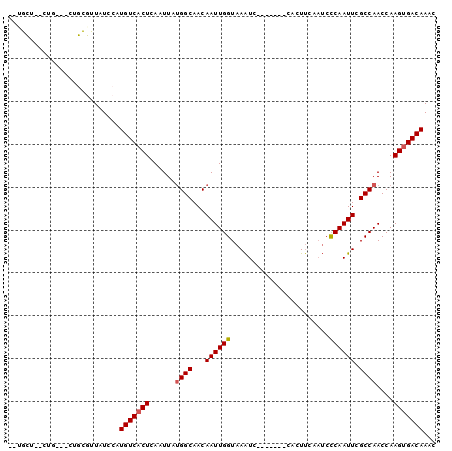
| Location | 1,544,087 – 1,544,183 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -14.39 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1544087 96 - 27905053 UCUGCU--CUG--GCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUCCCAAUCCCACUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC ...((.--...--...))........(((((((......((((...((((((.......................)))))).))))....)))))))... ( -17.30) >DroVir_CAF1 55436 87 - 1 CUUGGC--CUU----UCUCUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUC-------CACUUCAAUCUCAAUUCGCCAACCAAGUGACAAAC ......--...----...........(((((((......((....)).((((..(((.-------.............)))..))))...)))))))... ( -14.44) >DroEre_CAF1 45016 87 - 1 --UGCU--CUU--GCUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUC-------CACUUCCAUCCCAAUUCGCCAACCAAGGGACAAAC --((((--(((--(..(((.......(((((........)))))..((((((......-------..........)))))))))....)))))).))... ( -15.09) >DroWil_CAF1 46386 89 - 1 --AGCC--CUGGGUCUGUGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUC-------CACUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC --....--.(((((.......)))))(((((((......((((...((((((......-------..........)))))).))))....)))))))... ( -19.09) >DroYak_CAF1 41026 73 - 1 --------------------UAUCCAUGUCACUCAAUUGCGGCAACAAUUGGUAAGUC-------CACUCUAUACCCAAUUCGCCAACCAAGUGACAAAC --------------------......(((((((.......(((...((((((((((..-------....)).)).)))))).))).....)))))))... ( -15.90) >DroAna_CAF1 43477 91 - 1 CCCGCUCGCUC--CCCGCGCUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUC-------CACUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC ...((.((...--..)).))......(((((((......((((...((((((......-------..........)))))).))))....)))))))... ( -18.89) >consensus __UGCU__CUG___CUGCGUUAUCCAUGUCACUCAAUUAUGGCAACAAUUGGUAAAUC_______CACUUCAAUCCCAAUUCGCCAACCAAGUGACAAAC ..........................(((((((......((((...((((((.......................)))))).))))....)))))))... (-14.39 = -14.58 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:39 2006