
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,313,646 – 11,313,744 |
| Length | 98 |
| Max. P | 0.968235 |

| Location | 11,313,646 – 11,313,744 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.85 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
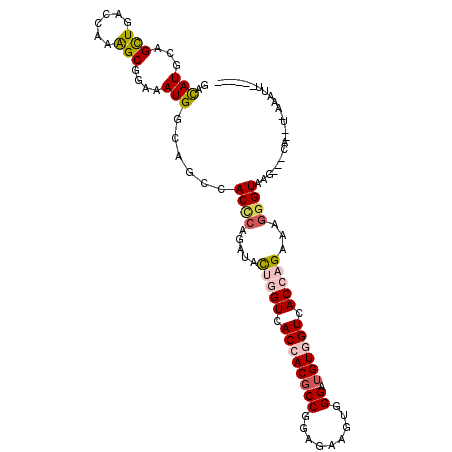
>3R_DroMel_CAF1 11313646 98 + 27905053 GAUAUACAGCUGACCAAGGCGGAAAUGGCUGCCACCCAGAUACUAGUCACCACGCCGGAGAAAUGGGAUGUGGUAACCAGAAAGGGUAAU---AA--U-AAUAU------ .................(((((......)))))((((.(((....)))((((((((.........)).)))))).........))))...---..--.-.....------ ( -24.10) >DroPse_CAF1 53206 99 + 1 GACAUGCAGCUGACCAAGGCGGAAAUGGCAGCCACCCAGAUAUUGGUAACCACGCCAGAGAAGUGGGAUGUGGUCACCAGAAAGGGUAAG---CA--UUAACUU------ ...((((.((((.(((.........))))))).((((.....(((((.((((((((.........)).)))))).)))))...))))..)---))--)......------ ( -31.30) >DroWil_CAF1 59716 101 + 1 GAUAUGCAGCUGACAAAAGCGGAGAUGGCGGCCACUCAAAUUCUCGUGACGACGCCGGAAAAGUGGGAUGUGGUCACACGAAAGGGUAGAUGCAA--U-AAAAA------ ....(((((((......))).(((.(((...))))))....(((((((((.(((((.(.....).)).))).))))).(....).).))))))).--.-.....------ ( -26.60) >DroMoj_CAF1 59596 106 + 1 GACAUGCAGCUGACCAAAGCAGAGAUGACGGCCACGCAGAUACUGGUCACCACGCCCGAAAAGUGGGAUGUUGUGACCAGAAAGGGUAAG---UCAAU-AAAUUGGCUCA (((.(((.(((......))).....((.((....))))....((((((((.(((((((.....)))).))).)))))))).....))).)---))...-........... ( -32.20) >DroAna_CAF1 49269 92 + 1 GACAUGCAGUUGACCAAAGCGGAGAUGACAGCCACCCAGAUUUUGGUUACCACCCCGGAGAAAUGGGAUGUGGUCACCCGAAAAGGUUAG---C---------U------ .......((((((((....(((.(.((((.((..((((..((((((........))))))...))))..)).))))))))....))))))---)---------)------ ( -29.90) >DroPer_CAF1 53364 99 + 1 GACAUGCAGCUGACCAAGGCGGAAAUGGCAGCCACCCAGAUAUUGGUGACCACGCCAGAGAAGUGGGAUGUGGUCACCAGAAAGGGUAAG---CA--UUAACUU------ ...((((.((((.(((.........))))))).((((.....((((((((((((((.........)).))))))))))))...))))..)---))--)......------ ( -37.50) >consensus GACAUGCAGCUGACCAAAGCGGAAAUGGCAGCCACCCAGAUACUGGUCACCACGCCGGAGAAGUGGGAUGUGGUCACCAGAAAGGGUAAG___CA__U_AAAUU______ ..(((...(((......)))....)))......((((.....(((((.((((((((.........)).)))))).)))))...))))....................... (-16.32 = -16.85 + 0.53)

| Location | 11,313,646 – 11,313,744 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.23 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
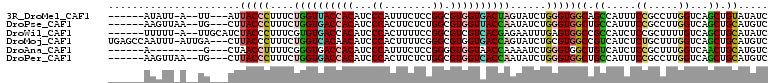
>3R_DroMel_CAF1 11313646 98 - 27905053 ------AUAUU-A--UU---AUUACCCUUUCUGGUUACCACAUCCCAUUUCUCCGGCGUGGUGACUAGUAUCUGGGUGGCAGCCAUUUCCGCCUUGGUCAGCUGUAUAUC ------.....-.--..---..(((((...(((((((((((...((........)).))))))))))).....)))))(((((.....((.....))...)))))..... ( -31.30) >DroPse_CAF1 53206 99 - 1 ------AAGUUAA--UG---CUUACCCUUUCUGGUGACCACAUCCCACUUCUCUGGCGUGGUUACCAAUAUCUGGGUGGCUGCCAUUUCCGCCUUGGUCAGCUGCAUGUC ------......(--((---(..((((....((((((((((...(((......))).))))))))))......))))((((((((.........))).)))))))))... ( -36.40) >DroWil_CAF1 59716 101 - 1 ------UUUUU-A--UUGCAUCUACCCUUUCGUGUGACCACAUCCCACUUUUCCGGCGUCGUCACGAGAAUUUGAGUGGCCGCCAUCUCCGCUUUUGUCAGCUGCAUAUC ------.....-.--.((((.((......(((((((((......((........)).)))).)))))((....((((((.........))))))...)))).)))).... ( -20.30) >DroMoj_CAF1 59596 106 - 1 UGAGCCAAUUU-AUUGA---CUUACCCUUUCUGGUCACAACAUCCCACUUUUCGGGCGUGGUGACCAGUAUCUGCGUGGCCGUCAUCUCUGCUUUGGUCAGCUGCAUGUC ((((.(((...-.))).---))))......((((((((.((..(((.......))).)).))))))))....((((((((((............))))))..)))).... ( -28.40) >DroAna_CAF1 49269 92 - 1 ------A---------G---CUAACCUUUUCGGGUGACCACAUCCCAUUUCUCCGGGGUGGUAACCAAAAUCUGGGUGGCUGUCAUCUCCGCUUUGGUCAACUGCAUGUC ------.---------(---(..((((....))))(((((((((((........))))))....(((.....)))((((..(....).))))..)))))....))..... ( -25.60) >DroPer_CAF1 53364 99 - 1 ------AAGUUAA--UG---CUUACCCUUUCUGGUGACCACAUCCCACUUCUCUGGCGUGGUCACCAAUAUCUGGGUGGCUGCCAUUUCCGCCUUGGUCAGCUGCAUGUC ------......(--((---(..((((....((((((((((...(((......))).))))))))))......))))((((((((.........))).)))))))))... ( -38.60) >consensus ______AAGUU_A__UG___CUUACCCUUUCUGGUGACCACAUCCCACUUCUCCGGCGUGGUCACCAAUAUCUGGGUGGCUGCCAUCUCCGCCUUGGUCAGCUGCAUGUC ......................(((((....((((((((((...((........)).))))))))))......)))))((.((.....((.....))...)).))..... (-20.28 = -20.23 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:23 2006