
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,308,724 – 11,308,822 |
| Length | 98 |
| Max. P | 0.848895 |

| Location | 11,308,724 – 11,308,822 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -15.30 |
| Energy contribution | -14.69 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11308724 98 + 27905053 G---------CUAAUCCAACGCAGCAAACGAGCGGCAAUCGAAAG-CCCAAUUCGAA--CGGUUAGC-UUUGGAUUGGCCAAGU----GCCGGCUAAUUGGAUUAG--GGCAAUUAG .---------(((((((((.((.((......))((((..(....)-.((((((((((--(.....).-)))))))))).....)----))).))...)))))))))--......... ( -30.40) >DroVir_CAF1 54104 115 + 1 GCUGUUAACACUAAUCCGACGCAGCAAAUUUGCAGCAAUUGGGAA-ACAAAUUUCAAAUUUAGCUAC-UUUGGAUUGGCCAAGUGUGUGCCGGAUAAUUGGAUUAGCUGGCAAUUAG ((((((((..(((((((((.(.((((((((((....((((.(...-.).)))).))))))).))).)-.)))))))))((((...(((.....))).)))).))))).)))...... ( -29.40) >DroPse_CAF1 47692 101 + 1 C---------CUAAUCCGACGCGACAAAUUUGCAGCAAUCGAAAG-CCCAAGUGGAAAUCCAACAGCUUUUGGAUUGGCCAAGU----GCAUCCUAAUUGGAUUAG--GGCAAUUAG (---------(((((((((.((((.....)))).(((..(....)-......(((.(((((((......)))))))..)))..)----)).......)))))))))--)........ ( -30.40) >DroMoj_CAF1 54568 103 + 1 G---------CUAAUCCGACACAACAAAUUUACCGCAAUUGGGAAGACGAAUUUAAAAUUGAGCGAC-UUUGGAUUGGCCAAGU----GACGGAUAAUUGGAUUAGCUAGCAAUUAG (---------(((((((((..........((((((((.((((.....(.((((((((.((....)).-)))))))).))))).)----).))).))))))))))))).......... ( -23.90) >DroAna_CAF1 44017 98 + 1 C---------CUAAUCCGACGCAGCAAAU-UGCGGCAAUCGAGGG-CCCAAUUCGAA-UCCGCCAAC-UUUGGAUUGGCCAAGU----GCCAUGUAAUUGGAUUAG--GGCAAUUAG (---------(((((((((.(((((....-.))((((......((-((.((((((((-.........-))))))))))))...)----))).)))..)))))))))--)........ ( -35.70) >DroPer_CAF1 47803 101 + 1 C---------CUAAUCCGACGCGGCAAAUUUGCAGCAAUCGAAAG-CCCAAGUGGAAAUCCAAAAGCUUUUGGAUUGGCCAAGU----GCAUCCUAAUUGGAUUAG--GGCAAUUAG (---------(((((((((.((.(((....))).)).........-..((..(((.((((((((....))))))))..)))..)----)........)))))))))--)........ ( -32.30) >consensus C_________CUAAUCCGACGCAGCAAAUUUGCAGCAAUCGAAAG_CCCAAUUCGAAAUCCAACAAC_UUUGGAUUGGCCAAGU____GCCGGCUAAUUGGAUUAG__GGCAAUUAG ..........(((((((((.((((.....))))..(((((.(((........(((....)))......))).)))))....................)))))))))........... (-15.30 = -14.69 + -0.61)



| Location | 11,308,724 – 11,308,822 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.25 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.00 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11308724 98 - 27905053 CUAAUUGCC--CUAAUCCAAUUAGCCGGC----ACUUGGCCAAUCCAAA-GCUAACCG--UUCGAAUUGGG-CUUUCGAUUGCCGCUCGUUUGCUGCGUUGGAUUAG---------C .........--((((((((((((((..((----...((((.((((.(((-((...(((--.......))))-)))).))))))))...))..)))).))))))))))---------. ( -32.60) >DroVir_CAF1 54104 115 - 1 CUAAUUGCCAGCUAAUCCAAUUAUCCGGCACACACUUGGCCAAUCCAAA-GUAGCUAAAUUUGAAAUUUGU-UUCCCAAUUGCUGCAAAUUUGCUGCGUCGGAUUAGUGUUAACAGC ..........((((((((........(((.((....)))))........-(((((.(((((((....(((.-....)))......))))))))))))...))))))))......... ( -28.40) >DroPse_CAF1 47692 101 - 1 CUAAUUGCC--CUAAUCCAAUUAGGAUGC----ACUUGGCCAAUCCAAAAGCUGUUGGAUUUCCACUUGGG-CUUUCGAUUGCUGCAAAUUUGUCGCGUCGGAUUAG---------G (((((((((--(..((((.....))))..----...(((..(((((((......))))))).)))...)))-)...((((.((.(((....))).))))))))))))---------. ( -30.00) >DroMoj_CAF1 54568 103 - 1 CUAAUUGCUAGCUAAUCCAAUUAUCCGUC----ACUUGGCCAAUCCAAA-GUCGCUCAAUUUUAAAUUCGUCUUCCCAAUUGCGGUAAAUUUGUUGUGUCGGAUUAG---------C ..........((((((((...........----..((((.....)))).-(.(((.(((.((((...((((..........)))))))).)))..))).))))))))---------) ( -19.60) >DroAna_CAF1 44017 98 - 1 CUAAUUGCC--CUAAUCCAAUUACAUGGC----ACUUGGCCAAUCCAAA-GUUGGCGGA-UUCGAAUUGGG-CCCUCGAUUGCCGCA-AUUUGCUGCGUCGGAUUAG---------G ........(--(((((((.......((((----.....)))).......-...((((.(-..(((((((((-(........))).))-))))).).)))))))))))---------) ( -32.50) >DroPer_CAF1 47803 101 - 1 CUAAUUGCC--CUAAUCCAAUUAGGAUGC----ACUUGGCCAAUCCAAAAGCUUUUGGAUUUCCACUUGGG-CUUUCGAUUGCUGCAAAUUUGCCGCGUCGGAUUAG---------G (((((((((--(..((((.....))))..----...(((..((((((((....)))))))).)))...)))-)...((((.((.(((....))).))))))))))))---------. ( -32.30) >consensus CUAAUUGCC__CUAAUCCAAUUAGCCGGC____ACUUGGCCAAUCCAAA_GCUGCUGGAUUUCAAAUUGGG_CUCUCGAUUGCUGCAAAUUUGCUGCGUCGGAUUAG_________C ...........(((((((.................((((.....)))).................................((.((......)).))...))))))).......... (-13.50 = -13.00 + -0.50)

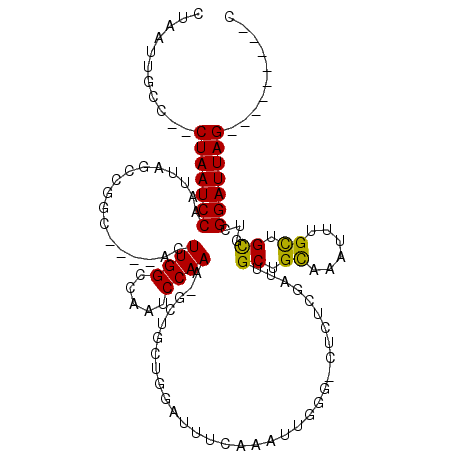
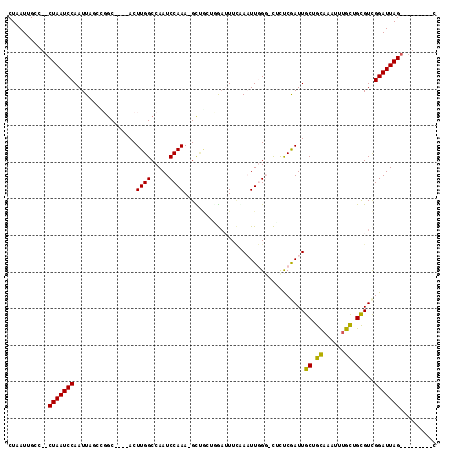
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:20 2006