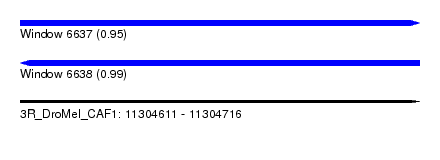
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,304,611 – 11,304,716 |
| Length | 105 |
| Max. P | 0.987805 |

| Location | 11,304,611 – 11,304,716 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -27.55 |
| Consensus MFE | -16.18 |
| Energy contribution | -17.35 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11304611 105 + 27905053 GCAAAAAUCAAACUCAAUUACUGCCGCUGUCAAUCAGCGGCAGGAAUCGAAUUGU-GAAUCGC---CGGCAUCCAAAU------CGGAAUAGGAAUAGAGAGGCAUGCGUAAGAG (((....(((...((.(((.(((((((((.....))))))))).))).))....)-))...((---(..(.(((....------.......)))...)...))).)))....... ( -30.10) >DroVir_CAF1 49748 81 + 1 GCAAAAAUCAAACUCAAUUACGUGCGCUGUCAAUCAGCGGCAGGAAUCGAAUUGG-GAAUCGG---------------------------------UCAAAGACAUGCGUAAGAG (((.........(((((((.(.(((((((.....)))).))).).....))))))-).....(---------------------------------((...))).)))....... ( -19.00) >DroGri_CAF1 45082 82 + 1 GCAAAAAUCAAAGUCAAUUACGUGCGCUGUCAAUCAGCGGCAGGAAUCGAACUCAAAAAUCGG---------------------------------ACAAAGACAUGCGUAAGAG (((....((...(((.....(.(((((((.....)))).))).)...(((.........))))---------------------------------))...))..)))....... ( -15.20) >DroSec_CAF1 39202 105 + 1 GCAAAAAUCAAACUCAAUUACUGCCGCUGUCAAUCAGCGGCAGGAAUCGAAUUGU-GAAUCGC---CGGCAUCCAAAU------CGGAAUACGAAUAGAGAGGCAUGCGUAAGAG (((....(((...((.(((.(((((((((.....))))))))).))).))....)-))...((---(....((....(------((.....)))...))..))).)))....... ( -32.10) >DroEre_CAF1 38540 102 + 1 GCAAAAAUCAAACUCAAUUACUGCCGCUGUCAAUCAGCGGCAGGAAUCGAAUUGG-GAAUC------GGCAUCCCAAU------CGGAAUCGGAAUAGAGAGGCAUGCGUAAGAG (((....((...........(((((((((.....)))))))))((.((..(((((-((...------....)))))))------..)).))........))....)))....... ( -33.90) >DroYak_CAF1 40072 114 + 1 GCAAAAAUCAAACUCAAUUACUGCCGCUGUCAAUCAGCGGCAGGAAUCGAAUUGG-GAAUCUCCAUCGCCAUCCAAAUCGGAAUCGGAAUCGGAAUAGAGAGCCAUGCGUAAGAG ............(((((((.(((((((((.....)))))))))......))))))-)...(((...(((..(((.....)))...((..((......))...))..)))...))) ( -35.00) >consensus GCAAAAAUCAAACUCAAUUACUGCCGCUGUCAAUCAGCGGCAGGAAUCGAAUUGG_GAAUCGC____GGCAUCCAAAU______CGGAAU_GGAAUAGAGAGGCAUGCGUAAGAG (((.................(((((((((.....)))))))))((.((........)).))............................................)))....... (-16.18 = -17.35 + 1.17)

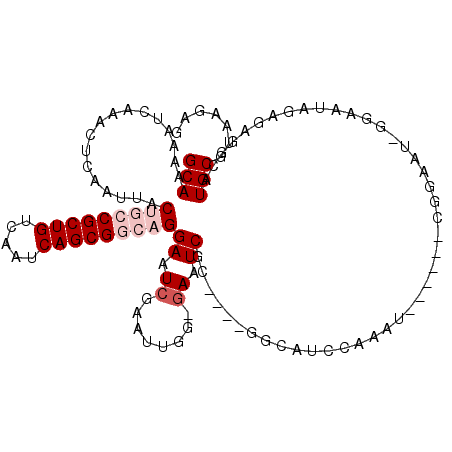
| Location | 11,304,611 – 11,304,716 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -20.17 |
| Energy contribution | -21.20 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11304611 105 - 27905053 CUCUUACGCAUGCCUCUCUAUUCCUAUUCCG------AUUUGGAUGCCG---GCGAUUC-ACAAUUCGAUUCCUGCCGCUGAUUGACAGCGGCAGUAAUUGAGUUUGAUUUUUGC ......(((..((.(((..(((........)------))..))).))..---)))..((-(.(((((((((.(((((((((.....))))))))).))))))))))))....... ( -35.40) >DroVir_CAF1 49748 81 - 1 CUCUUACGCAUGUCUUUGA---------------------------------CCGAUUC-CCAAUUCGAUUCCUGCCGCUGAUUGACAGCGCACGUAAUUGAGUUUGAUUUUUGC .......(((.(((...))---------------------------------)......-(.((((((((((.(((.((((.....))))))).).))))))))).).....))) ( -16.20) >DroGri_CAF1 45082 82 - 1 CUCUUACGCAUGUCUUUGU---------------------------------CCGAUUUUUGAGUUCGAUUCCUGCCGCUGAUUGACAGCGCACGUAAUUGACUUUGAUUUUUGC .......(((.(((...((---------------------------------((((.........))).....(((.((((.....))))))).......)))...)))...))) ( -15.30) >DroSec_CAF1 39202 105 - 1 CUCUUACGCAUGCCUCUCUAUUCGUAUUCCG------AUUUGGAUGCCG---GCGAUUC-ACAAUUCGAUUCCUGCCGCUGAUUGACAGCGGCAGUAAUUGAGUUUGAUUUUUGC ......(((..((.(((....(((.....))------)...))).))..---)))..((-(.(((((((((.(((((((((.....))))))))).))))))))))))....... ( -36.90) >DroEre_CAF1 38540 102 - 1 CUCUUACGCAUGCCUCUCUAUUCCGAUUCCG------AUUGGGAUGCC------GAUUC-CCAAUUCGAUUCCUGCCGCUGAUUGACAGCGGCAGUAAUUGAGUUUGAUUUUUGC .......(((....((..(((((((((....------)))))))))..------))...-(.(((((((((.(((((((((.....))))))))).))))))))).).....))) ( -34.80) >DroYak_CAF1 40072 114 - 1 CUCUUACGCAUGGCUCUCUAUUCCGAUUCCGAUUCCGAUUUGGAUGGCGAUGGAGAUUC-CCAAUUCGAUUCCUGCCGCUGAUUGACAGCGGCAGUAAUUGAGUUUGAUUUUUGC .......((((((.(((((((((((..(((((.......)))))))).))))))))...-)))((((((((.(((((((((.....))))))))).))))))))........))) ( -43.00) >consensus CUCUUACGCAUGCCUCUCUAUUCC_AUUCCG______AUUUGGAUGCC____GCGAUUC_CCAAUUCGAUUCCUGCCGCUGAUUGACAGCGGCAGUAAUUGAGUUUGAUUUUUGC ..............................................................(((((((((.(((((((((.....))))))))).))))))))).......... (-20.17 = -21.20 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:17 2006