
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,302,220 – 11,302,319 |
| Length | 99 |
| Max. P | 0.962497 |

| Location | 11,302,220 – 11,302,319 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -23.27 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
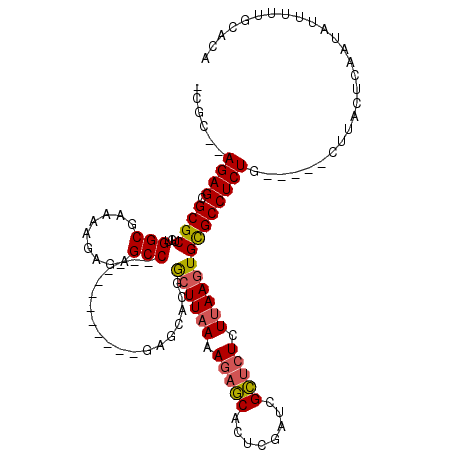
>3R_DroMel_CAF1 11302220 99 + 27905053 -CGC--AGAGAGCGCUGUGGCGAAAAGAGAGCC------------GAGCGCGGCUUAAAAGAGCACUCGAACGCUCUCUUAAGUGCGCCUCUG-----CUUACUCAAUAUUUUUGCACA -.((--(((((.(((....)))....((((((.------------(((((((.(((((.(((((........))))).))))))))).))).)-----))..)))....)))))))... ( -38.20) >DroSec_CAF1 36939 99 + 1 -CGC--AGAGCGCGCUGUGGCGAAAAGAGAGCC------------GAGCACGGCUUAAAAGAGCACUCGAUCGCUCUCUUAAGUGCGCCUCUG-----CUUACUCAAUAUUUCUGCACA -.((--((((((((((........(((((((((------------(((....((((....)))).))))...)))))))).)))))).)))))-----).................... ( -37.90) >DroSim_CAF1 37735 99 + 1 -CGC--AGAGCGCGCUGUGGCGAAAAGAGAGCC------------GAGCACGGCUUAAAAGAGCACUCGAUCGCUCUCUUAAGUGCGCCUCUG-----CUUACUCAAUAUUUUUGCACA -.((--((((((((((........(((((((((------------(((....((((....)))).))))...)))))))).)))))).)))))-----).................... ( -37.90) >DroYak_CAF1 37202 119 + 1 UCAAAGAGAGCGCGCUCUGGCGAAAAGAAAGCCAAACUCGCGGCCGAGCGCGACUUCAAAACUCAUUCCGUGGAUCUCUUAAAUGUGCCUCUGUGCUUCUGGCUCGAUAUUUUCGCACA ...((((((.(((((((.(((.....(..((.....))..).))))))))))..............((....))))))))...(((((....(.((.....)).)((.....))))))) ( -31.10) >consensus _CGC__AGAGCGCGCUGUGGCGAAAAGAGAGCC____________GAGCACGGCUUAAAAGAGCACUCGAUCGCUCUCUUAAGUGCGCCUCUG_____CUUACUCAAUAUUUUUGCACA ......((((.((((...(((.........)))...................((((((.(((((........))))).))))))))))))))........................... (-23.27 = -23.52 + 0.25)

| Location | 11,302,220 – 11,302,319 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -26.10 |
| Energy contribution | -26.23 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
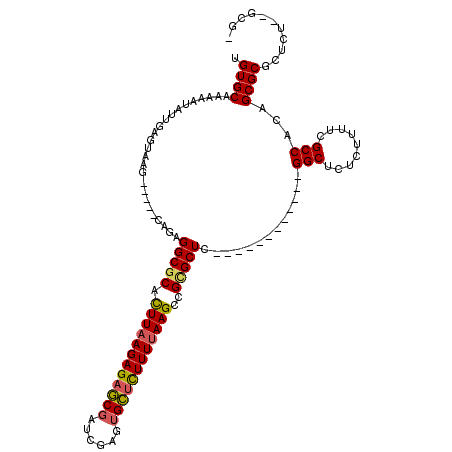
>3R_DroMel_CAF1 11302220 99 - 27905053 UGUGCAAAAAUAUUGAGUAAG-----CAGAGGCGCACUUAAGAGAGCGUUCGAGUGCUCUUUUAAGCCGCGCUC------------GGCUCUCUUUUCGCCACAGCGCUCUCU--GCG- ....................(-----(((((((((....((((((((...(((((((...........))))))------------))))))))).........)))).))))--)).- ( -38.62) >DroSec_CAF1 36939 99 - 1 UGUGCAGAAAUAUUGAGUAAG-----CAGAGGCGCACUUAAGAGAGCGAUCGAGUGCUCUUUUAAGCCGUGCUC------------GGCUCUCUUUUCGCCACAGCGCGCUCU--GCG- ....................(-----(((((((((..(((((((((((......)))))))))))((((....)------------)))...............)))).))))--)).- ( -38.10) >DroSim_CAF1 37735 99 - 1 UGUGCAAAAAUAUUGAGUAAG-----CAGAGGCGCACUUAAGAGAGCGAUCGAGUGCUCUUUUAAGCCGUGCUC------------GGCUCUCUUUUCGCCACAGCGCGCUCU--GCG- ....................(-----(((((((((..(((((((((((......)))))))))))((((....)------------)))...............)))).))))--)).- ( -38.10) >DroYak_CAF1 37202 119 - 1 UGUGCGAAAAUAUCGAGCCAGAAGCACAGAGGCACAUUUAAGAGAUCCACGGAAUGAGUUUUGAAGUCGCGCUCGGCCGCGAGUUUGGCUUUCUUUUCGCCAGAGCGCGCUCUCUUUGA ((.(((((((....((((((((.((...(((((((.(((((((..((........)).))))))))).)).)))....))...))))))))..)))))))))(((....)))....... ( -33.30) >consensus UGUGCAAAAAUAUUGAGUAAG_____CAGAGGCGCACUUAAGAGAGCGAUCGAGUGCUCUUUUAAGCCGCGCUC____________GGCUCUCUUUUCGCCACAGCGCGCUCU__GCG_ .((((.........................(((((.((((((((((((......))))))))))))..))))).............(((.........)))...))))........... (-26.10 = -26.23 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:15 2006