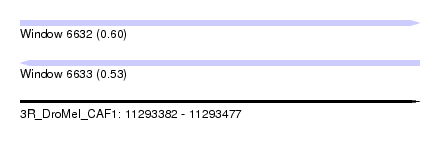
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,293,382 – 11,293,477 |
| Length | 95 |
| Max. P | 0.601263 |

| Location | 11,293,382 – 11,293,477 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11293382 95 + 27905053 UCUGUUAUUGUUAUUGUU---------------AUUGCUCUGGCUGUGCAUUUGAAUGCACAUCGAUUUGGCGGCCAGCAGGCUCUAAAUUAUUAAUUCCAUAAUAUCUA ..((((((.((((...((---------------(..(((((((((((((((....)))).((......))))))))))..)))..))).....))))...)))))).... ( -22.20) >DroSec_CAF1 28129 95 + 1 UCUGUUAUUCUUAUUUUU---------------AUUGCUCUGGCUGUGCAUUUGGAUGCAAAUCGAUUUGGCGGCCAGCAGGCUCUAAAUUAUUAAUUCCAUAAUAUCUA ..((..(((..((..(((---------------(..(((((((((((.((..(.(((....))).)..))))))))))..)))..)))).))..)))..))......... ( -22.30) >DroSim_CAF1 28816 95 + 1 UCUGUAAUUGUUAUUGUU---------------AUUGCUCUGGCUGUGCAUUUGGAUGCAAAUCGAUUUGGCGGCCAGCAGGCUUUAAAUUAUUAAUUCCAUAAUAUCUA ..((.(((((.((...((---------------(..(((((((((((.((..(.(((....))).)..))))))))))..)))..)))..)).))))).))......... ( -23.50) >DroEre_CAF1 27282 101 + 1 UCUGUUAUUAUUAUUGUUAUUGUG---------AUUGCUCUGGCUGUGCAUUUUGAUGCAAAUCGUUUUGGCGGCCAGCGGGCUCUAAAUUAUGAGUUCCAUAAUCUAAA ..................((((((---------......((((((((.((...((((....))))...)))))))))).((((((........))))))))))))..... ( -25.50) >DroYak_CAF1 27779 110 + 1 UCUGUGAUUGUUAUUGUUAUUGUUAUUGCUGUUAUUGCUCUGGCUGUGCAUUUUGAUGCAAAUCGUUUUGGCGGCCAGCAGGCUCUGAAUUAUUAGUUCCAUAAUCCAUA ..((.((((((...............(((((...(((((..(((..(((((....)))))....)))..))))).)))))((..((((....))))..)))))))))).. ( -24.50) >consensus UCUGUUAUUGUUAUUGUU_______________AUUGCUCUGGCUGUGCAUUUGGAUGCAAAUCGAUUUGGCGGCCAGCAGGCUCUAAAUUAUUAAUUCCAUAAUAUCUA ....................................(((((((((((((((....))))....(.....)))))))))..)))........................... (-16.36 = -16.36 + -0.00)



| Location | 11,293,382 – 11,293,477 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 85.12 |
| Mean single sequence MFE | -18.08 |
| Consensus MFE | -13.66 |
| Energy contribution | -13.66 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11293382 95 - 27905053 UAGAUAUUAUGGAAUUAAUAAUUUAGAGCCUGCUGGCCGCCAAAUCGAUGUGCAUUCAAAUGCACAGCCAGAGCAAU---------------AACAAUAACAAUAACAGA (((((((((......)))).)))))..((...(((((((......)).(((((((....)))))))))))).))...---------------.................. ( -19.30) >DroSec_CAF1 28129 95 - 1 UAGAUAUUAUGGAAUUAAUAAUUUAGAGCCUGCUGGCCGCCAAAUCGAUUUGCAUCCAAAUGCACAGCCAGAGCAAU---------------AAAAAUAAGAAUAACAGA ....(((((......))))).((((..((...(((((((......))...(((((....)))))..))))).))..)---------------)))............... ( -15.30) >DroSim_CAF1 28816 95 - 1 UAGAUAUUAUGGAAUUAAUAAUUUAAAGCCUGCUGGCCGCCAAAUCGAUUUGCAUCCAAAUGCACAGCCAGAGCAAU---------------AACAAUAACAAUUACAGA (((((((((......)))).)))))..((...(((((((......))...(((((....)))))..))))).))...---------------.................. ( -14.50) >DroEre_CAF1 27282 101 - 1 UUUAGAUUAUGGAACUCAUAAUUUAGAGCCCGCUGGCCGCCAAAACGAUUUGCAUCAAAAUGCACAGCCAGAGCAAU---------CACAAUAACAAUAAUAAUAACAGA (((((((((((.....)))))))))))((...(((((((......))...(((((....)))))..))))).))...---------........................ ( -21.50) >DroYak_CAF1 27779 110 - 1 UAUGGAUUAUGGAACUAAUAAUUCAGAGCCUGCUGGCCGCCAAAACGAUUUGCAUCAAAAUGCACAGCCAGAGCAAUAACAGCAAUAACAAUAACAAUAACAAUCACAGA ..((((((((.......))))))))....((((((((((......))...(((((....)))))..))))).((.......)).......................))). ( -19.80) >consensus UAGAUAUUAUGGAAUUAAUAAUUUAGAGCCUGCUGGCCGCCAAAUCGAUUUGCAUCCAAAUGCACAGCCAGAGCAAU_______________AACAAUAACAAUAACAGA ...........................((...(((((((......))...(((((....)))))..))))).)).................................... (-13.66 = -13.66 + 0.00)

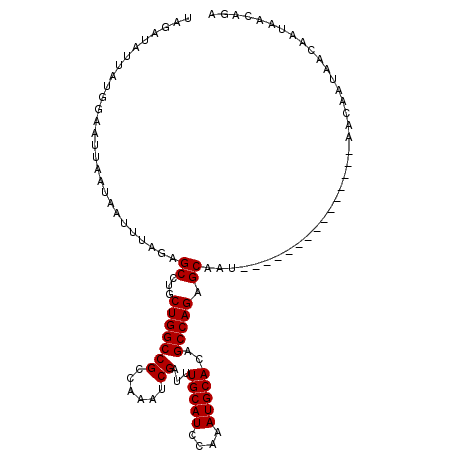

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:13 2006