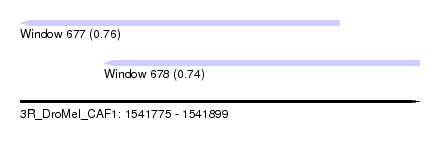
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,541,775 – 1,541,899 |
| Length | 124 |
| Max. P | 0.759460 |

| Location | 1,541,775 – 1,541,874 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -11.14 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
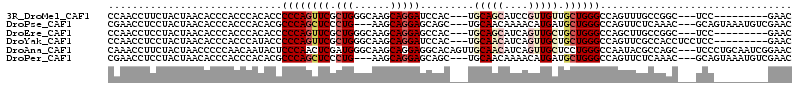
>3R_DroMel_CAF1 1541775 99 - 27905053 CCAACCUUCUACUAACACCCACCCACACCCCCAGUUCGCUGGGCAAGCAGGAUCCAC---UGCAGCAUCCGUUGUUGCUGGGCCAGUUUGCCGGC---UCC---------GAAC .................................(((((.(.(((((((.((.((((.---.((((((.....)))))))))))).))))))).).---..)---------)))) ( -29.20) >DroPse_CAF1 42610 105 - 1 CGAACCUCCUACUAACACCCACCCACACGCCCAGCUCCCUG---AAGCAGGAGCAGC---UGCAACAAAACAUGAUGCUGGGCCAGUUCUCAAAC---GCAGUAAAUGUCGAAC (((......((((...............(((((((((((((---...)))).((...---.))..........)).)))))))..(((....)))---..))))....)))... ( -23.50) >DroEre_CAF1 42664 99 - 1 CCAACCUCCUACUAACACCCACCCACACCCCCAGUUCGCUGGGCAAGCAGGAGCCAC---UGCAGCAUCAGUUGCUGCUGGGCCAGCUUGCCGGC---UCC---------GAAC .............................(((((....)))))......((((((.(---.((((((.....)))))).)(((......))))))---)))---------.... ( -35.70) >DroYak_CAF1 38730 102 - 1 CCAACCUCCUACUAACACCCACCCAUACCCCCAGUUCGCUGGGCAAGCAGGAUCCAC---UGCAACAUCAGUUGCUGCUGGGCCAGUUCGCCACCUCCUCC---------GAAC .....................(((.....(((((....)))))..(((((....)..---.(((((....))))))))))))...(((((..........)---------)))) ( -23.80) >DroAna_CAF1 41230 111 - 1 CAAACCUUCUACUAACCCCCAACAAUACUCCCAACUCGAUGGGCAAGCAGGAGGCACAGUUGCAACAUCAGUUGCUCCUGGGCCAAUACGCCAGC---UCCCUGCAAUCGGAAC ..................((.........((((......))))...(((((..((.(((..(((((....)))))..)))(((......))).))---..)))))....))... ( -35.60) >DroPer_CAF1 44130 105 - 1 CGAACCUCCUACUAACACCCACCCACACGCCCAGCUCCCUG---AAGCAGGAGCAGC---UGCAACAAAACAUGAUGCUGGGCCAGUUCUCAAAC---GCAGUAAAUGUCGAAC (((......((((...............(((((((((((((---...)))).((...---.))..........)).)))))))..(((....)))---..))))....)))... ( -23.50) >consensus CCAACCUCCUACUAACACCCACCCACACCCCCAGCUCGCUGGGCAAGCAGGAGCCAC___UGCAACAUCAGUUGCUGCUGGGCCAGUUCGCCAGC___UCC_________GAAC .............................((((((((.(((......))))).........(((((....))))).))))))................................ (-11.14 = -12.28 + 1.14)
| Location | 1,541,801 – 1,541,899 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -11.31 |
| Energy contribution | -13.00 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1541801 98 - 27905053 CAGCAGCAACAUUUGCUGACCACCUCCAACCUUCUACUAACACCCACCCACACCCCCAGUUCGCUGGGCAAGCAGGAUCCAC---UGCAGCAUCCGUUGUU ....((((((...((((.....................................(((((....)))))...((((......)---)))))))...)))))) ( -24.20) >DroPse_CAF1 42645 95 - 1 CAGCAGCAACACUUGUUAGCAACAUCGAACCUCCUACUAACACCCACCCACACGCCCAGCUCCCUG---AAGCAGGAGCAGC---UGCAACAAAACAUGAU ..(((((......((((((.................))))))................(((((...---.....))))).))---)))............. ( -21.53) >DroEre_CAF1 42690 98 - 1 CACCAGCAACAUUUGCUGACCACCUCCAACCUCCUACUAACACCCACCCACACCCCCAGUUCGCUGGGCAAGCAGGAGCCAC---UGCAGCAUCAGUUGCU ....((((((...((((.....................................(((((....)))))...((((......)---)))))))...)))))) ( -26.60) >DroYak_CAF1 38759 98 - 1 CAGCAGCAACAUUUGCUGACCACCUCCAACCUCCUACUAACACCCACCCAUACCCCCAGUUCGCUGGGCAAGCAGGAUCCAC---UGCAACAUCAGUUGCU ((((((......))))))........((((........................(((((....)))))...((((......)---))).......)))).. ( -23.10) >DroAna_CAF1 41265 101 - 1 CAACAGCAACAUUUGCUGGCAACAUCAAACCUUCUACUAACCCCCAACAAUACUCCCAACUCGAUGGGCAAGCAGGAGGCACAGUUGCAACAUCAGUUGCU ....((((((...((...(((((......((((((.((....((((.(..............).))))..)).))))))....)))))..))...)))))) ( -24.34) >DroPer_CAF1 44165 95 - 1 CAGCAGCAACACUUGUUGGCAACAUCGAACCUCCUACUAACACCCACCCACACGCCCAGCUCCCUG---AAGCAGGAGCAGC---UGCAACAAAACAUGAU ..(((((.....(((.((....)).)))..............................(((((...---.....))))).))---)))............. ( -22.20) >consensus CAGCAGCAACAUUUGCUGACAACAUCCAACCUCCUACUAACACCCACCCACACCCCCAGCUCGCUGGGCAAGCAGGAGCCAC___UGCAACAUCAGUUGCU ...(((((.....)))))............(((((.((................(((((....)))))..)).)))))........(((((....))))). (-11.31 = -13.00 + 1.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:37 2006