
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,288,877 – 11,288,987 |
| Length | 110 |
| Max. P | 0.990317 |

| Location | 11,288,877 – 11,288,987 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -14.16 |
| Energy contribution | -15.08 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
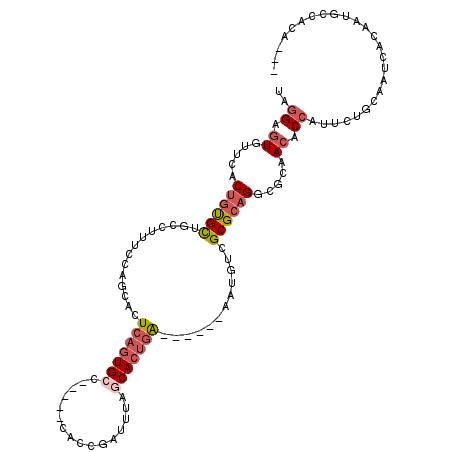
>3R_DroMel_CAF1 11288877 110 + 27905053 UAGGAGUGUUCACUGUGCUGCCUUUCCAGCACUCAGUGCC-----CACCGAUUUAGCACUGAGUCGUAAAUGUCGCGCAGGCGCAACACCAUUCUGCAAUCACAAUGCCACAAUC ..((.((((((.((((((.((.(((...(.(((((((((.-----..........))))))))))..))).)).))))))..).)))))((((.((....)).))))))...... ( -31.70) >DroSec_CAF1 23656 104 + 1 UGGGAGUGUUCACUGCGUAGCCUUUGCAGCACUCAGUGCC-----CACCGAUUUAGCACUGA------AAUGUCGCGCAGGCGCAACACCAUUCUGCAAUCACAAUGCCACAAUC ..((((((.....(((...((((.(((.(((.(((((((.-----..........)))))))------..))).))).)))))))....))))))(((.......)))....... ( -28.90) >DroSim_CAF1 24293 104 + 1 UGGGAGUGUUCACUGCGCUGCCUUUCCAGCACUCAGUGCC-----CACCGAUUUAGCACUGA------AAUGUCGCGCAGGCGCAACACCAUUCUGCAAUCACAAUGCCACAAUC (((..((((((.(((((((((.......))).(((((((.-----..........)))))))------......))))))..).)))))((((.((....)).)))))))..... ( -28.90) >DroEre_CAF1 22724 97 + 1 UAGGAGUUUUCACUGUGCUGCCUUUCCCGCACUCAGUGAC-----CACCGAUUCAGCACUGA------AAUGUCGCGCAGGCGCAACUCCAUUCUGCAAUCACAAUGC------- ..((((((....(((((((((.......))).((((((..-----...........))))))------......))))))....))))))..................------- ( -25.72) >DroYak_CAF1 23134 95 + 1 UAGGAGUUUUCACUGUGU------UACAGCGCUCAGUGCC-----CACCGAUUUAGCACUGA------AAUGUCGCGCAGGCGCAACUCCAUUCUGCAAUCACAAUGCCACA--- ..((((((....((((((------.(((....(((((((.-----..........)))))))------..))).))))))....)))))).....(((.......)))....--- ( -30.20) >DroMoj_CAF1 29914 96 + 1 --UGACUAUUUACACUGCAUGUUUCACAGUAAUCUUUGGCAAUGGCUAUGAUCAAACACUCG------AUAGCUGCGCAGACGCAACGCCAACAUGCGAUUCAC----------- --.............(((((((..............((((...((((((((........)).------))))))(((....)))...)))))))))))......----------- ( -21.43) >consensus UAGGAGUGUUCACUGUGCUGCCUUUCCAGCACUCAGUGCC_____CACCGAUUUAGCACUGA______AAUGUCGCGCAGGCGCAACACCAUUCUGCAAUCACAAUGCCACA___ ..((.((.....((((((..............(((((((................)))))))............)))))).....)).))......................... (-14.16 = -15.08 + 0.92)

| Location | 11,288,877 – 11,288,987 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -22.82 |
| Energy contribution | -22.93 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11288877 110 - 27905053 GAUUGUGGCAUUGUGAUUGCAGAAUGGUGUUGCGCCUGCGCGACAUUUACGACUCAGUGCUAAAUCGGUG-----GGCACUGAGUGCUGGAAAGGCAGCACAGUGAACACUCCUA ((.(((..(((((((.((((.....(((((((((....)))))))))....((((((((((.........-----)))))))))).((....))))))))))))).))).))... ( -44.30) >DroSec_CAF1 23656 104 - 1 GAUUGUGGCAUUGUGAUUGCAGAAUGGUGUUGCGCCUGCGCGACAUU------UCAGUGCUAAAUCGGUG-----GGCACUGAGUGCUGCAAAGGCUACGCAGUGAACACUCCCA ((((.((((((((............(((((((((....)))))))))------.))))))))))))((((-----..(((((.(((((.....))).)).)))))..)))).... ( -40.32) >DroSim_CAF1 24293 104 - 1 GAUUGUGGCAUUGUGAUUGCAGAAUGGUGUUGCGCCUGCGCGACAUU------UCAGUGCUAAAUCGGUG-----GGCACUGAGUGCUGGAAAGGCAGCGCAGUGAACACUCCCA ((((.((((((((............(((((((((....)))))))))------.))))))))))))((((-----..((((..((((((......))))))))))..)))).... ( -41.12) >DroEre_CAF1 22724 97 - 1 -------GCAUUGUGAUUGCAGAAUGGAGUUGCGCCUGCGCGACAUU------UCAGUGCUGAAUCGGUG-----GUCACUGAGUGCGGGAAAGGCAGCACAGUGAAAACUCCUA -------.(((((((.((((........((((((....)))))).((------((..(((....(((((.-----...)))))..))).)))).))))))))))).......... ( -32.90) >DroYak_CAF1 23134 95 - 1 ---UGUGGCAUUGUGAUUGCAGAAUGGAGUUGCGCCUGCGCGACAUU------UCAGUGCUAAAUCGGUG-----GGCACUGAGCGCUGUA------ACACAGUGAAAACUCCUA ---.((..(((((((.((((((......((((((....))))))..(------((((((((.........-----)))))))))..)))))------))))))))...))..... ( -37.60) >DroMoj_CAF1 29914 96 - 1 -----------GUGAAUCGCAUGUUGGCGUUGCGUCUGCGCAGCUAU------CGAGUGUUUGAUCAUAGCCAUUGCCAAAGAUUACUGUGAAACAUGCAGUGUAAAUAGUCA-- -----------.(((...(((...((((((((((....)))))).((------(((....)))))....)))).))).......((((((.......)))))).......)))-- ( -24.40) >consensus ___UGUGGCAUUGUGAUUGCAGAAUGGUGUUGCGCCUGCGCGACAUU______UCAGUGCUAAAUCGGUG_____GGCACUGAGUGCUGGAAAGGCAGCACAGUGAACACUCCUA ........(((((((.............((((((....))))))..........((((((....((((((.......)))))))))))).........))))))).......... (-22.82 = -22.93 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:11 2006