| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,281,938 – 11,282,035 |
| Length | 97 |
| Max. P | 0.529572 |

| Location | 11,281,938 – 11,282,035 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -24.78 |
| Consensus MFE | -12.92 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11281938 97 + 27905053 CGAAAAUAGUCAGACGUUUAUGUGAUUUUUCUU-CGCCACACACGCAGACC---UGAAUGCU------UAGAGUUACUCACAGGUAAAUGGUCAUGGUCUGACUCAU .......((((((((......((((.......)-)))..........((((---....((((------(.(((...)))..)))))...))))...))))))))... ( -23.80) >DroPse_CAF1 22471 96 + 1 UAAAAAUAGUCAGACAUUUAUAUGAUUUUCCCUGUGCCACACA--CAGACC---AAAAUUCC------CAAUUCUAUUCACAGGUGAAUGGUCAAUGGCUGACUCAC .......((((((..........((((((..(((((.....))--)))...---)))))).(------((...(((((((....)))))))....)))))))))... ( -22.20) >DroSim_CAF1 16556 97 + 1 CAAAAAUAGUCAGACGUUUAUGUGAUUUUCCUU-CGCCACACACGCAGACC---UGAAUGCU------UAAAGUUACUCACAGGUAAAUGGUCAUGGUCUGACUCAU .......((((((((......((((.......)-)))..........((((---....((((------(............)))))...))))...))))))))... ( -21.60) >DroEre_CAF1 15765 97 + 1 CGAAAAUAGUCAGACGUUUGUGUGGUUUUCCUU-CGCCACACACGCAGACC---UGAAUGCU------UAAGGUUACUCACAGGUAAAUGGUCAUGGUCUGACUCAU .......((((((((((.((((((((.......-.)))))))).)).((((---....((((------(.((....))...)))))...))))...))))))))... ( -34.30) >DroAna_CAF1 16556 105 + 1 UAAAAAUAGUCAGACAUUUAUGUGAUUUUCCCUGUGCCACACA--CAGACCAACAGAAUGCCAGAACGCAGAGUUACUCAGAGGUAAAUGGCCAAGGUCUGACUCAU .......((((((((.(((..(((((((...(((((.....))--)))..........(((......))))))))))..)))(((.....)))...))))))))... ( -26.50) >DroPer_CAF1 22574 96 + 1 UAAAAAUAGUCAGACAUUUAUAUGAUUUUCCCUGUGCCACACA--CAGACC---AAAAUUCC------CAGUUCUAUUCGCAGGUGAAUGGUCAAUGGCUGAUUCAC .......((((((..........((((((..(((((.....))--)))...---)))))).(------((...(((((((....)))))))....)))))))))... ( -20.30) >consensus CAAAAAUAGUCAGACAUUUAUGUGAUUUUCCCU_CGCCACACA__CAGACC___AGAAUGCC______CAAAGUUACUCACAGGUAAAUGGUCAAGGUCUGACUCAU .......((((((((.....((((.............))))......((((......................(((((....)))))..))))...))))))))... (-12.92 = -13.50 + 0.59)



| Location | 11,281,938 – 11,282,035 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.94 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -18.61 |
| Energy contribution | -17.70 |
| Covariance contribution | -0.91 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11281938 97 - 27905053 AUGAGUCAGACCAUGACCAUUUACCUGUGAGUAACUCUA------AGCAUUCA---GGUCUGCGUGUGUGGCG-AAGAAAAAUCACAUAAACGUCUGACUAUUUUCG ...((((((((...........(((((...((.......------.))...))---)))..(..(((((((..-........)))))))..)))))))))....... ( -23.20) >DroPse_CAF1 22471 96 - 1 GUGAGUCAGCCAUUGACCAUUCACCUGUGAAUAGAAUUG------GGAAUUUU---GGUCUG--UGUGUGGCACAGGGAAAAUCAUAUAAAUGUCUGACUAUUUUUA ...((((((.((((((((((((.((.((.......)).)------))))...)---))))((--((.....))))..............)))).))))))....... ( -23.40) >DroSim_CAF1 16556 97 - 1 AUGAGUCAGACCAUGACCAUUUACCUGUGAGUAACUUUA------AGCAUUCA---GGUCUGCGUGUGUGGCG-AAGGAAAAUCACAUAAACGUCUGACUAUUUUUG ...((((((((...........(((((...((.......------.))...))---)))..(..((((((((.-...)....)))))))..)))))))))....... ( -24.00) >DroEre_CAF1 15765 97 - 1 AUGAGUCAGACCAUGACCAUUUACCUGUGAGUAACCUUA------AGCAUUCA---GGUCUGCGUGUGUGGCG-AAGGAAAACCACACAAACGUCUGACUAUUUUCG ...((((((((...((((.........((((....))))------........---)))).(..((((((((.-...)....)))))))..)))))))))....... ( -28.33) >DroAna_CAF1 16556 105 - 1 AUGAGUCAGACCUUGGCCAUUUACCUCUGAGUAACUCUGCGUUCUGGCAUUCUGUUGGUCUG--UGUGUGGCACAGGGAAAAUCACAUAAAUGUCUGACUAUUUUUA ...(((((((((((((((((.(((....((.((((..(((......)))....)))).)).)--)).))))).))))).......((....)))))))))....... ( -31.30) >DroPer_CAF1 22574 96 - 1 GUGAAUCAGCCAUUGACCAUUCACCUGCGAAUAGAACUG------GGAAUUUU---GGUCUG--UGUGUGGCACAGGGAAAAUCAUAUAAAUGUCUGACUAUUUUUA .....((((.((((((((((((.((..(.....)....)------))))...)---))))((--((.....))))..............)))).))))......... ( -18.40) >consensus AUGAGUCAGACCAUGACCAUUUACCUGUGAGUAACUCUA______AGCAUUCA___GGUCUG__UGUGUGGCA_AAGGAAAAUCACAUAAACGUCUGACUAUUUUUA ...((((((((...(((((((((....)))))....((.......)).........))))....(((((((...........)))))))...))))))))....... (-18.61 = -17.70 + -0.91)

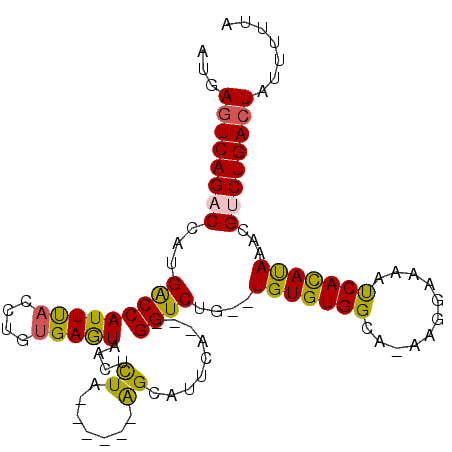

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:06 2006