
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,274,710 – 11,274,807 |
| Length | 97 |
| Max. P | 0.920405 |

| Location | 11,274,710 – 11,274,807 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -26.78 |
| Energy contribution | -27.45 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11274710 97 + 27905053 GUGAGACGGAGACCAUCGAGUGCGGAUCGUCCUCGACGUAACCGGAGUAGAAGCAGCCUAGGUGGCGCUGCUCCGCCUCCGCCUCCGUCUCCGGUCC (.(((((((((....(((((..((...))..)))))......(((((..(.((((((((....)).)))))).)..))))).))))))))))..... ( -44.70) >DroPse_CAF1 15077 95 + 1 GCGAAACGGCCACCGCCGAGUGGGAGUCGCCUUCGACGUAGCCCUUGUAGUAGCAGCCCAGAUAGUGCUGCUGCUCCUGGUCCU--GUUCCUGCUCC .......(((....)))(((..(((((((....)))).(((..((.(.(((((((((.(.....).))))))))).).))..))--).)))..))). ( -36.80) >DroSec_CAF1 9738 97 + 1 GUGAGACGGAGACCAUCGAGUGCGGAUCGUCCUCGACGUAGCCGGAGUAUAAGCAGCCCAGAUGGCGCUGUUCCGCCUCAGCCUCCGUCUCCGGUCC (.(((((((((....(((((..((...))..))))).(.((.((((......(((((((....)).))))))))).)))...))))))))))..... ( -39.60) >DroSim_CAF1 9661 97 + 1 GUGAGACGGAGACCAUCGAGUGCGGAUCGUCCUCGACGUAGCCGGAGUAGAAGCAGCCCAGGUGGCGCUGCUCCGCCUCCGCCUCCGUCUCCGGUCC (.(((((((((....(((((..((...))..)))))......(((((..(.((((((((....)).)))))).)..))))).))))))))))..... ( -47.20) >DroYak_CAF1 8447 91 + 1 GUGAGACGGAGACCAUCGAGUGCGGAUCGUCCUCGACGUAGCCGGAGUAGAAGCAGCCCAGGUGGCGCUGCUCCGCCUCC------GUCUCCGGUCC (.(((((((((....(((((..((...))..)))))......((((((((..((.((....)).)).)))))))).))))------))))))..... ( -42.40) >DroPer_CAF1 15194 95 + 1 GCGAAACGGCCACCGCCGAGUGGGAGUCGCCUUCGACGUAGCCCUUGUAGUAGCAGCCCAGAUAGUGCUGCUGCUCCUGGUCCU--GUUCCUGCUCC .......(((....)))(((..(((((((....)))).(((..((.(.(((((((((.(.....).))))))))).).))..))--).)))..))). ( -36.80) >consensus GUGAGACGGAGACCAUCGAGUGCGGAUCGUCCUCGACGUAGCCGGAGUAGAAGCAGCCCAGAUGGCGCUGCUCCGCCUCCGCCU__GUCUCCGGUCC ......(((((((..(((((.(((...))).))))).......((((..(.((((((((....)).)))))).)..))))......))))))).... (-26.78 = -27.45 + 0.67)


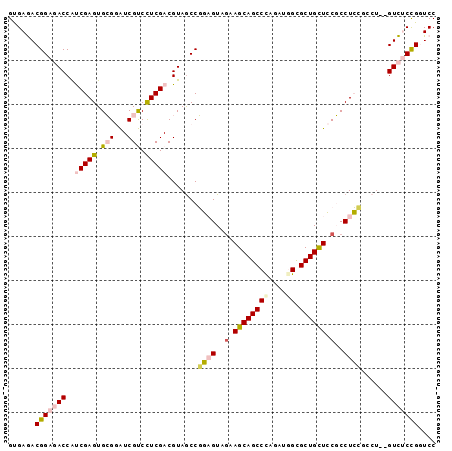
| Location | 11,274,710 – 11,274,807 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -46.08 |
| Consensus MFE | -32.28 |
| Energy contribution | -32.67 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11274710 97 - 27905053 GGACCGGAGACGGAGGCGGAGGCGGAGCAGCGCCACCUAGGCUGCUUCUACUCCGGUUACGUCGAGGACGAUCCGCACUCGAUGGUCUCCGUCUCAC ......(((((((((((((((..((((((((.........))))))))..)))).....(((((((..((...))..)))))))))))))))))).. ( -54.00) >DroPse_CAF1 15077 95 - 1 GGAGCAGGAAC--AGGACCAGGAGCAGCAGCACUAUCUGGGCUGCUACUACAAGGGCUACGUCGAAGGCGACUCCCACUCGGCGGUGGCCGUUUCGC ...((.(((((--.......(.((.((((((.((....)))))))).)).)...(((((((((((.((......))..))))).))))))))))))) ( -33.60) >DroSec_CAF1 9738 97 - 1 GGACCGGAGACGGAGGCUGAGGCGGAACAGCGCCAUCUGGGCUGCUUAUACUCCGGCUACGUCGAGGACGAUCCGCACUCGAUGGUCUCCGUCUCAC ......(((((((((((..((.((((..((((((.....))).))).....)))).)).(((((((..((...))..)))))))))))))))))).. ( -47.40) >DroSim_CAF1 9661 97 - 1 GGACCGGAGACGGAGGCGGAGGCGGAGCAGCGCCACCUGGGCUGCUUCUACUCCGGCUACGUCGAGGACGAUCCGCACUCGAUGGUCUCCGUCUCAC ......(((((((((((((((..((((((((.((....))))))))))..)))).....(((((((..((...))..)))))))))))))))))).. ( -57.80) >DroYak_CAF1 8447 91 - 1 GGACCGGAGAC------GGAGGCGGAGCAGCGCCACCUGGGCUGCUUCUACUCCGGCUACGUCGAGGACGAUCCGCACUCGAUGGUCUCCGUCUCAC ((((.((((((------((((..((((((((.((....))))))))))..)))).....(((((((..((...))..)))))))))))))))))... ( -50.10) >DroPer_CAF1 15194 95 - 1 GGAGCAGGAAC--AGGACCAGGAGCAGCAGCACUAUCUGGGCUGCUACUACAAGGGCUACGUCGAAGGCGACUCCCACUCGGCGGUGGCCGUUUCGC ...((.(((((--.......(.((.((((((.((....)))))))).)).)...(((((((((((.((......))..))))).))))))))))))) ( -33.60) >consensus GGACCGGAGAC__AGGCGGAGGCGGAGCAGCGCCACCUGGGCUGCUUCUACUCCGGCUACGUCGAGGACGAUCCGCACUCGAUGGUCUCCGUCUCAC ......(((((......((((..((((((((.((....))))))))))..))))((...(((((((...........)))))))....))))))).. (-32.28 = -32.67 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:03 2006