
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,244,094 – 11,244,184 |
| Length | 90 |
| Max. P | 0.998012 |

| Location | 11,244,094 – 11,244,184 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.25 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
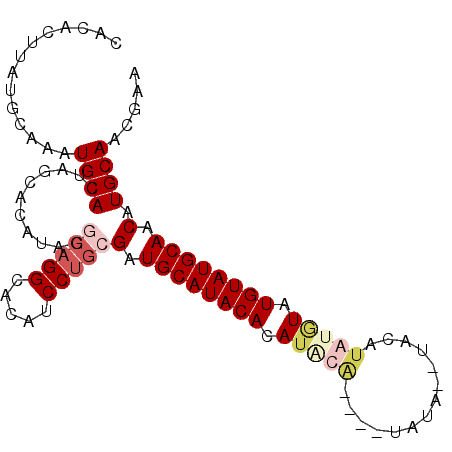
>3R_DroMel_CAF1 11244094 90 + 27905053 UUCGUUGCAUGUUGCAUACAUA----UGUA--UAUA----UGUAUGUGUAUGCAUCGCAGGAUAUGCCUGCCUAUGUGCUAGUGCAUUUUCAUAAGUGUG ......((((((((((((((((----((..--....----..))))))))))))..(((((.....)))))..))))))....((((((....)))))). ( -29.10) >DroSim_CAF1 3283 96 + 1 UUCGUUGCAUGUUGCAUACAUACAUAUGUAUGUGUA----UGUAUGUGUAUGCAUCGCAGGAUGUGCCUGCCUAUGUGCUAGUGCAUUUGCAUAAGUGUG ...(.(((((...(((((((((((((....))))))----)))))))..))))).)(((((.....)))))....(..((.((((....)))).))..). ( -36.20) >DroEre_CAF1 3302 86 + 1 UUCGUUGCAUGUUGCAUACAUACAUACAGA--GAUAUGUGCACAUGUGUAUGCAUCGCAGGAUGUGCC------------AGUGCAUUUGCAUAAGUGUG ......(((((.((((((((((..((((..--....))))....)))))))))).)((((.(((((..------------..)))))))))....)))). ( -23.20) >DroYak_CAF1 3278 94 + 1 UUCGUUGCAUGUUGCAUACAUAUUUACACA--CAUA----CAUAUGUGUAUGCAUCGCAGGAUGUGCCUGUCUAUGUGCUAGUGCAUUUGCAUGAGUGUG ......(((((.(((((((((((.......--....----...)))))))))))..(((((.....))))).)))))(((.((((....)))).)))... ( -26.74) >consensus UUCGUUGCAUGUUGCAUACAUACAUACAUA__UAUA____CAUAUGUGUAUGCAUCGCAGGAUGUGCCUGCCUAUGUGCUAGUGCAUUUGCAUAAGUGUG .....((((((.((((((((((......................)))))))))).)(((((.....)))))..........))))).............. (-19.62 = -20.25 + 0.62)


| Location | 11,244,094 – 11,244,184 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.58 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -14.38 |
| Energy contribution | -16.57 |
| Covariance contribution | 2.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11244094 90 - 27905053 CACACUUAUGAAAAUGCACUAGCACAUAGGCAGGCAUAUCCUGCGAUGCAUACACAUACA----UAUA--UACA----UAUGUAUGCAACAUGCAACGAA .....(((((....(((....))))))))(((((.....)))))(.(((((...((((((----(((.--...)----))))))))....))))).)... ( -23.50) >DroSim_CAF1 3283 96 - 1 CACACUUAUGCAAAUGCACUAGCACAUAGGCAGGCACAUCCUGCGAUGCAUACACAUACA----UACACAUACAUAUGUAUGUAUGCAACAUGCAACGAA ........((((.((((((((.....)))(((((.....)))))..)))))...((((((----((((........)))))))))).....))))..... ( -28.20) >DroEre_CAF1 3302 86 - 1 CACACUUAUGCAAAUGCACU------------GGCACAUCCUGCGAUGCAUACACAUGUGCACAUAUC--UCUGUAUGUAUGUAUGCAACAUGCAACGAA ..............((((.(------------((((.....)))..((((((((.(((((((......--..))))))).)))))))).))))))..... ( -22.50) >DroYak_CAF1 3278 94 - 1 CACACUCAUGCAAAUGCACUAGCACAUAGACAGGCACAUCCUGCGAUGCAUACACAUAUG----UAUG--UGUGUAAAUAUGUAUGCAACAUGCAACGAA .((((.((((((.(((.....(((....(.((((.....)))))..))).....))).))----))))--.)))).....((((((...))))))..... ( -23.70) >consensus CACACUUAUGCAAAUGCACUAGCACAUAGGCAGGCACAUCCUGCGAUGCAUACACAUACA____UAUA__UACAUAUGUAUGUAUGCAACAUGCAACGAA ..............((((...........(((((.....)))))(.((((((((.(((((..............))))).)))))))).).))))..... (-14.38 = -16.57 + 2.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:52 2006