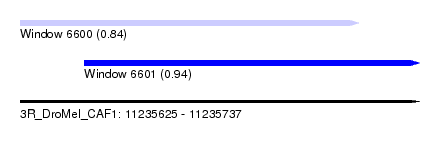
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,235,625 – 11,235,737 |
| Length | 112 |
| Max. P | 0.939388 |

| Location | 11,235,625 – 11,235,720 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.68 |
| Mean single sequence MFE | -15.61 |
| Consensus MFE | -10.44 |
| Energy contribution | -10.47 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11235625 95 + 27905053 ---------------CCAUU---CCCCACC-CCUGUC---CUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAGGCACAGCCACAAACGCAGCCA ---------------.....---.......-......---.....((((((((((.........((((....))))..........)))))....(((...)))......))))).. ( -19.71) >DroSec_CAF1 6765 98 + 1 GCCGUAGCCU---UCCCAUU---CCUCUCC-CCUGUCCUCCUCCUUCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAGUCACAGC------------CA ((((((((..---.......---...(...-...).............))))))..........((((....))))..((((((.........))))))..))------------.. ( -12.21) >DroSim_CAF1 8044 113 + 1 GCCGUAGCCGUCGUCCCACU---CCCCACC-CCUCUUCUCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAGUCACAGCCACAAACGCAGCCA ...((((.............---.......-............)))).((((((..........((((....))))..((((((.........))))))..........)))))).. ( -17.15) >DroEre_CAF1 7070 80 + 1 ---------------------------UCC-CCUU---UCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACA------GCCACAAACGCAACCA ---------------------------...-....---.......((((.(((((.........((((....))))..........)))))..))------)).............. ( -13.41) >DroYak_CAF1 8062 81 + 1 ---------------------------UCCACCCU---UCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACA------GCCACAAACGCAGCCA ---------------------------........---.....((((.((((((..(.......((((....))))...)..))))))((.....------)).......))))... ( -14.80) >DroAna_CAF1 6714 90 + 1 GUC-------------CACUUUUUUGCGCC-CCAAUGUUUGCGCUGCUGCUGCGCCCUUAA-AUUUUCAAUUGAAAUAUGACUGCAGUGCACACAUCCACAUCCA------------ ...-------------..............-...((((.((((((((.((...))......-..((((....)))).......)))))))).)))).........------------ ( -16.40) >consensus ________________CA_U___CC_CUCC_CCUGU__UCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAG_CACAGCCACAAACGCAGCCA ............................................(((.((((((..(.......((((....))))...)..))))))))).......................... (-10.44 = -10.47 + 0.03)



| Location | 11,235,643 – 11,235,737 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.41 |
| Mean single sequence MFE | -14.92 |
| Consensus MFE | -10.44 |
| Energy contribution | -10.47 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11235643 94 + 27905053 ---CUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAGGCACAGCCACAAACGCAGCCACACAACAACAUUCACAU----------- ---.....((((((((((.........((((....))))..........)))))....(((...)))......)))))...................----------- ( -19.71) >DroSec_CAF1 6795 85 + 1 CUCCUCCUUCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAGUCACAGC------------CACACAUCAACAUUCACAU----------- ...........((((............((((....))))..((((((.........))))))))))------------...................----------- ( -11.30) >DroSim_CAF1 8077 97 + 1 CUCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAGUCACAGCCACAAACGCAGCCACACAUCAACAUUCACAU----------- .......((..((((((..........((((....))))..((((((.........))))))..........))))))..))...............----------- ( -17.30) >DroEre_CAF1 7077 90 + 1 -UCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACA------GCCACAAACGCAACCACACAUCAACAUCCACAU----------- -.......((((.(((((.........((((....))))..........)))))..))------))...............................----------- ( -13.41) >DroYak_CAF1 8070 101 + 1 -UCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACA------GCCACAAACGCAGCCACACAUCAACAUCCACAUCCACAUCCAUA -.....((((.((((((..(.......((((....))))...)..))))))((.....------)).......))))............................... ( -14.80) >DroAna_CAF1 6737 81 + 1 UUUGCGCUGCUGCUGCGCCCUUAA-AUUUUCAAUUGAAAUAUGACUGCAGUGCACACAUCCACAUCCA--------------CAUCCACA-UCACAU----------- ..((((((((.((...))......-..((((....)))).......))))))))..............--------------........-......----------- ( -13.00) >consensus _UCCUCCUGCUGCUGCGCCCUUAAAAUUUUCAAUUGAAAUAUGACUGCAGCGCACACAG_CACAGCCACAAACGCAGCCACACAUCAACAUUCACAU___________ .......(((.((((((..(.......((((....))))...)..)))))))))...................................................... (-10.44 = -10.47 + 0.03)

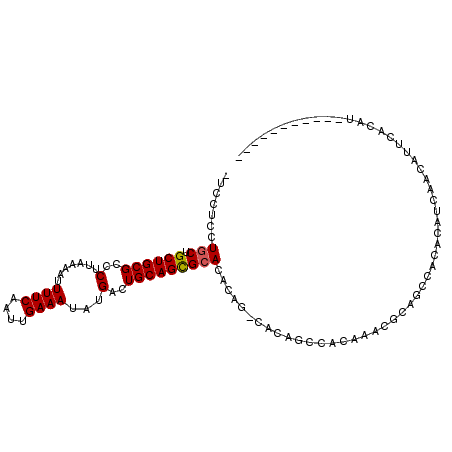
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:41 2006