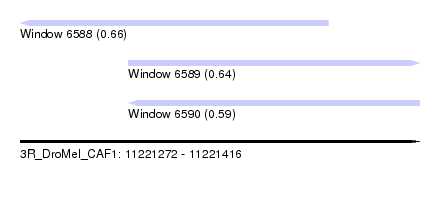
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,221,272 – 11,221,416 |
| Length | 144 |
| Max. P | 0.658402 |

| Location | 11,221,272 – 11,221,383 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11221272 111 - 27905053 AACGAAAUAUAGCCCUAUUUUAGCCCCGA---UUUCGGGCUUCUUGGGCUCUCUGAAU----UAUGUUCAGAUUUCGGUUCAUUACUUGCAUUUGCACAAGCUUUGGUAUGCAGUUAA ..(((((...(((((......(((((.(.---...))))))....))))).(((((((----...)))))))))))).....(((.((((((..((....))......)))))).))) ( -31.10) >DroSec_CAF1 23147 111 - 1 AACGAAAUAUAGCCCUAUUUCAGCCCCGA---UAUCGGGCUUCUUGGGCUCUCUGAAU----UAUGUUCAGAUUUCGGUACAUUACUUGCAUUUGCACAAGCUUUGGUAUGCAGUUAA ..(((((...(((((......(((((.(.---...))))))....))))).(((((((----...)))))))))))).....(((.((((((..((....))......)))))).))) ( -30.40) >DroSim_CAF1 23767 111 - 1 AACGAAAUAUAGCCCUAUUUUAGCCCCGA---UAUCGGGCUUCUUGGGCUCUCUGAAU----UAUGUUCAGAUUUCGGUACAUUACUUGCAUUUGCACAAGCUUUGGUAUGCAGUUAA ..(((((...(((((......(((((.(.---...))))))....))))).(((((((----...)))))))))))).....(((.((((((..((....))......)))))).))) ( -31.10) >DroEre_CAF1 28590 115 - 1 AACGAAAUAUAGCCCUAUUUGAACCCCGC---UUUCGGACUUCUUGGGUUCUCUGAAUUACUUAUGUUCAGAUAUCGGUUCAUUUCUUGCAUUUGCACAAGCUUUUGUAUGCAGUUAA ((((((((..((((......((((((...---.............))))))(((((((.......)))))))....)))).))))).(((((..((....))......)))))))).. ( -24.99) >DroYak_CAF1 24087 115 - 1 AACGAAAUAUAGCCCUAUUCGAACCCCGA---UUUCGGACUUCUUGGGUUCUCUGAAUUACUUAUGUUCAGAUUUCGGUUCAUUUCUUGCAUUUGCACAAGCUUUUGUAUGCAGUUGA ((((((((..((((......((((((.((---.........))..))))))(((((((.......)))))))....)))).))))).(((((..((....))......)))))))).. ( -26.40) >DroAna_CAF1 23739 115 - 1 UACGAAUUGUUGCCCUAUU-CAGCUCCAAAGUUUUGGGAGCUCUGGGCGUUUCUGAAUUGC--AUAUUCAGAGUUUUGUUCUUUAGUGGCAAUUGCAUAAGCAUUCGUAUGCAUUGUA ((((((..(((((((((..-.((((((.........))))))..(((((.((((((((...--..))))))))...)))))..))).))))))(((....)))))))))......... ( -35.70) >consensus AACGAAAUAUAGCCCUAUUUCAGCCCCGA___UUUCGGACUUCUUGGGCUCUCUGAAU____UAUGUUCAGAUUUCGGUUCAUUACUUGCAUUUGCACAAGCUUUGGUAUGCAGUUAA ..(((((...(((((......((.(((((.....))))).))...))))).(((((((.......)))))))))))).........((((((..((....))......)))))).... (-21.10 = -21.85 + 0.75)



| Location | 11,221,311 – 11,221,416 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643429 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11221311 105 + 27905053 ACCGAAAUCUGAACAUA----AUUCAGAGAGCCCAAGAAGCCCGAAA---UCGGGGCUAAAAUAGGGCUAUAUUUCGUUAGCCA-AACGAA--GAAGGUAAUAUUUAAU----UCAAUU (((....((((((....----.)))))).(((((....(((((....---...)))))......)))))...(((((((.....-))))))--)..)))..........----...... ( -28.80) >DroSec_CAF1 23186 105 + 1 ACCGAAAUCUGAACAUA----AUUCAGAGAGCCCAAGAAGCCCGAUA---UCGGGGCUGAAAUAGGGCUAUAUUUCGUUAGCCA-AACGAA--GAAGGUAAUAUUUAAU----UCAAUU (((....((((((....----.)))))).(((((....(((((....---...)))))......)))))...(((((((.....-))))))--)..)))..........----...... ( -29.90) >DroSim_CAF1 23806 105 + 1 ACCGAAAUCUGAACAUA----AUUCAGAGAGCCCAAGAAGCCCGAUA---UCGGGGCUAAAAUAGGGCUAUAUUUCGUUAGCCA-AACGAA--GAAGGUAAUAUUUAAU----UCAAUU (((....((((((....----.)))))).(((((....(((((....---...)))))......)))))...(((((((.....-))))))--)..)))..........----...... ( -28.80) >DroEre_CAF1 28629 109 + 1 ACCGAUAUCUGAACAUAAGUAAUUCAGAGAACCCAAGAAGUCCGAAA---GCGGGGUUCAAAUAGGGCUAUAUUUCGUUAGCCA-AACGAA--GAAGGUAAUAUUUAAU----UCAAUU (((....((((((.........))))))((((((.....((......---)).))))))......(((((........))))).-......--...)))..........----...... ( -23.60) >DroYak_CAF1 24126 110 + 1 ACCGAAAUCUGAACAUAAGUAAUUCAGAGAACCCAAGAAGUCCGAAA---UCGGGGUUCGAAUAGGGCUAUAUUUCGUUAGCCAAAACGAA--GAAGGUAAUAUUUAAU----UCAAUU ...(((.((((((.........))))))..(((...(((.((((...---.)))).)))......(((((........)))))........--...))).........)----)).... ( -24.50) >DroAna_CAF1 23778 115 + 1 ACAAAACUCUGAAUAU--GCAAUUCAGAAACGCCCAGAGCUCCCAAAACUUUGGAGCUG-AAUAGGGCAACAAUUCGUAAGACA-AGCGAAAAAAAGGUAAUAUUGAAUAAACACACUU .(((...(((((((..--...))))))).((((((..((((((.........)))))).-....)))).....(((((......-.)))))......))....)))............. ( -25.40) >consensus ACCGAAAUCUGAACAUA____AUUCAGAGAGCCCAAGAAGCCCGAAA___UCGGGGCUAAAAUAGGGCUAUAUUUCGUUAGCCA_AACGAA__GAAGGUAAUAUUUAAU____UCAAUU .......((((((.........))))))............(((((.....)))))....(((((..(((....((((((......)))))).....)))..)))))............. (-16.74 = -16.93 + 0.20)


| Location | 11,221,311 – 11,221,416 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.89 |
| Mean single sequence MFE | -25.57 |
| Consensus MFE | -17.92 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11221311 105 - 27905053 AAUUGA----AUUAAAUAUUACCUUC--UUCGUU-UGGCUAACGAAAUAUAGCCCUAUUUUAGCCCCGA---UUUCGGGCUUCUUGGGCUCUCUGAAU----UAUGUUCAGAUUUCGGU ......----..........(((...--((((((-.....))))))....(((((......(((((.(.---...))))))....))))).(((((((----...)))))))....))) ( -27.70) >DroSec_CAF1 23186 105 - 1 AAUUGA----AUUAAAUAUUACCUUC--UUCGUU-UGGCUAACGAAAUAUAGCCCUAUUUCAGCCCCGA---UAUCGGGCUUCUUGGGCUCUCUGAAU----UAUGUUCAGAUUUCGGU ......----..........(((...--((((((-.....))))))....(((((......(((((.(.---...))))))....))))).(((((((----...)))))))....))) ( -27.00) >DroSim_CAF1 23806 105 - 1 AAUUGA----AUUAAAUAUUACCUUC--UUCGUU-UGGCUAACGAAAUAUAGCCCUAUUUUAGCCCCGA---UAUCGGGCUUCUUGGGCUCUCUGAAU----UAUGUUCAGAUUUCGGU ......----..........(((...--((((((-.....))))))....(((((......(((((.(.---...))))))....))))).(((((((----...)))))))....))) ( -27.70) >DroEre_CAF1 28629 109 - 1 AAUUGA----AUUAAAUAUUACCUUC--UUCGUU-UGGCUAACGAAAUAUAGCCCUAUUUGAACCCCGC---UUUCGGACUUCUUGGGUUCUCUGAAUUACUUAUGUUCAGAUAUCGGU ......----..........(((...--((((((-.....))))))....(((((.....(((..(((.---...)))..)))..))))).(((((((.......)))))))....))) ( -22.10) >DroYak_CAF1 24126 110 - 1 AAUUGA----AUUAAAUAUUACCUUC--UUCGUUUUGGCUAACGAAAUAUAGCCCUAUUCGAACCCCGA---UUUCGGACUUCUUGGGUUCUCUGAAUUACUUAUGUUCAGAUUUCGGU ......----................--((((..(.(((((........)))))..)..))))..((((---....(((((.....)))))(((((((.......)))))))..)))). ( -23.20) >DroAna_CAF1 23778 115 - 1 AAGUGUGUUUAUUCAAUAUUACCUUUUUUUCGCU-UGUCUUACGAAUUGUUGCCCUAUU-CAGCUCCAAAGUUUUGGGAGCUCUGGGCGUUUCUGAAUUGC--AUAUUCAGAGUUUUGU (((((.........................))))-).....(((((.....((((....-.((((((.........))))))..))))..((((((((...--..)))))))).))))) ( -25.71) >consensus AAUUGA____AUUAAAUAUUACCUUC__UUCGUU_UGGCUAACGAAAUAUAGCCCUAUUUCAGCCCCGA___UUUCGGACUUCUUGGGCUCUCUGAAU____UAUGUUCAGAUUUCGGU ....................(((.....((((((......))))))....(((((......((.(((((.....))))).))...))))).(((((((.......)))))))....))) (-17.92 = -18.75 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:30 2006