
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,213,171 – 11,213,315 |
| Length | 144 |
| Max. P | 0.997489 |

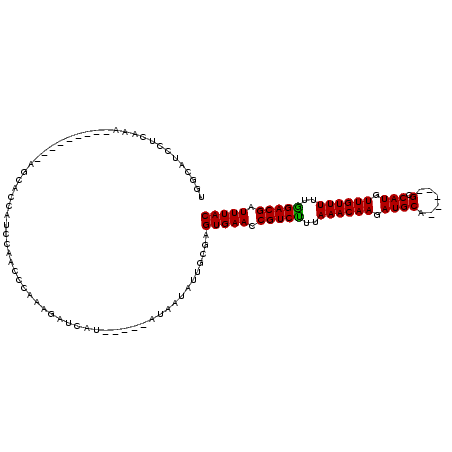
| Location | 11,213,171 – 11,213,275 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -19.61 |
| Energy contribution | -19.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.983126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11213171 104 + 27905053 UGGCAUCCUUUGA---------AGCACCAUCCAACCCCAAGAUCAA-----AUACUAAUGCCAGUGAACCGUCUUUAAACAAGAUGCACACAUGUCAUGUUGUUUUUAGACGAUUUAC ((((((..(((((---------....................))))-----).....))))))(((((.(((((..((((((.(((.((....))))).))))))..))))).))))) ( -26.65) >DroEre_CAF1 20363 108 + 1 UGGCAUCCUCAAACGAAGGAUUGGCACCAUCCAACCCGAAGAUCGU-----AUAAUAUGGCGAGUGAACCGUCCUUAAACAAGAUGCA-----GCCAUGUUGUUUUUGGACGAUUUAC .(.(((......((((.((.((((......)))).)).....))))-----.....))).)..(((((.(((((..((((((.(((..-----..))).))))))..))))).))))) ( -29.70) >DroYak_CAF1 15862 104 + 1 UGGCAUCCUGAAA---------AGCACCACCCAACCCAAUGAUCAUAAUAUAUAUUAUUGCGAGUGAACCGUCUUUAAACAAGAUGCA-----GCCAUGUUGUUUUUGGACGAUUUAC (((....((....---------))..))).....(.(((((((.(((...)))))))))).).(((((.(((((..((((((.(((..-----..))).))))))..))))).))))) ( -22.30) >consensus UGGCAUCCUCAAA_________AGCACCAUCCAACCCAAAGAUCAU_____AUAAUAUUGCGAGUGAACCGUCUUUAAACAAGAUGCA_____GCCAUGUUGUUUUUGGACGAUUUAC ...............................................................(((((.(((((..((((((.((((......).))).))))))..))))).))))) (-19.61 = -19.17 + -0.44)


| Location | 11,213,171 – 11,213,275 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.21 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -20.07 |
| Energy contribution | -19.74 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11213171 104 - 27905053 GUAAAUCGUCUAAAAACAACAUGACAUGUGUGCAUCUUGUUUAAAGACGGUUCACUGGCAUUAGUAU-----UUGAUCUUGGGGUUGGAUGGUGCU---------UCAAAGGAUGCCA ((.((((((((..((((((.(((((....)).))).))))))..)))))))).)).(((((((.(((-----(..(((....)))..)))).).((---------....)))))))). ( -30.70) >DroEre_CAF1 20363 108 - 1 GUAAAUCGUCCAAAAACAACAUGGC-----UGCAUCUUGUUUAAGGACGGUUCACUCGCCAUAUUAU-----ACGAUCUUCGGGUUGGAUGGUGCCAAUCCUUCGUUUGAGGAUGCCA ((.((((((((..((((((.(((..-----..))).))))))..)))))))).)).((((((.....-----.(((((....))))).))))))...((((((.....)))))).... ( -35.70) >DroYak_CAF1 15862 104 - 1 GUAAAUCGUCCAAAAACAACAUGGC-----UGCAUCUUGUUUAAAGACGGUUCACUCGCAAUAAUAUAUAUUAUGAUCAUUGGGUUGGGUGGUGCU---------UUUCAGGAUGCCA .......(.(((.........))))-----.((((((((...((((((.(..((((((..(((((....)))))......)))).))..).)).))---------)).)))))))).. ( -22.00) >consensus GUAAAUCGUCCAAAAACAACAUGGC_____UGCAUCUUGUUUAAAGACGGUUCACUCGCAAUAAUAU_____AUGAUCUUCGGGUUGGAUGGUGCU_________UUUAAGGAUGCCA ((.((((((((..((((((.(((.........))).))))))..)))))))).))..................(((((....)))))..(((((((..............)).))))) (-20.07 = -19.74 + -0.33)



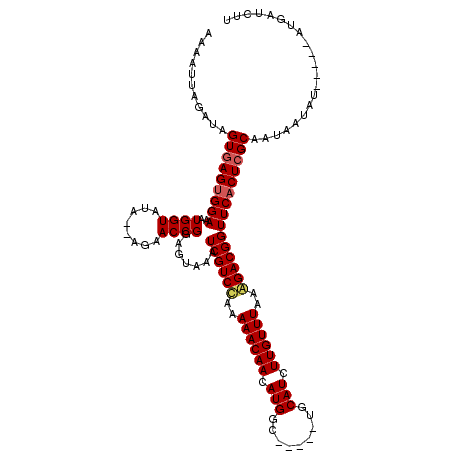
| Location | 11,213,200 – 11,213,315 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11213200 115 + 27905053 AAGAUCAA-----AUACUAAUGCCAGUGAACCGUCUUUAAACAAGAUGCACACAUGUCAUGUUGUUUUUAGACGAUUUACUCCGGUUUCCAUUUACCAUUUCCACUCACUAUUUAACUUU ........-----...........(((((..(((((..((((((.(((.((....))))).))))))..))))).........(((........)))........))))).......... ( -24.80) >DroEre_CAF1 20401 108 + 1 AAGAUCGU-----AUAAUAUGGCGAGUGAACCGUCCUUAAACAAGAUGCA-----GCCAUGUUGUUUUUGGACGAUUUACUUCGGUUCG--AAUACCAUUUCCGCUCACUAUCUAAUUUU ........-----.....((((.(((((((((((((..((((((.(((..-----..))).))))))..))))((......)))))))(--((......))).)))).))))........ ( -30.70) >DroYak_CAF1 15891 113 + 1 AUGAUCAUAAUAUAUAUUAUUGCGAGUGAACCGUCUUUAAACAAGAUGCA-----GCCAUGUUGUUUUUGGACGAUUUACUCCGGUUCU--UAUACCAUUUCCACUCACUAUCUAAUUUU ............((((..((((.(((((((.(((((..((((((.(((..-----..))).))))))..))))).)))))))))))...--))))......................... ( -27.00) >consensus AAGAUCAU_____AUAAUAUUGCGAGUGAACCGUCUUUAAACAAGAUGCA_____GCCAUGUUGUUUUUGGACGAUUUACUCCGGUUCC__UAUACCAUUUCCACUCACUAUCUAAUUUU .......................(((((((.(((((..((((((.((((......).))).))))))..))))).))))))).(((........)))....................... (-25.39 = -25.17 + -0.22)


| Location | 11,213,200 – 11,213,315 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.18 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -23.35 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11213200 115 - 27905053 AAAGUUAAAUAGUGAGUGGAAAUGGUAAAUGGAAACCGGAGUAAAUCGUCUAAAAACAACAUGACAUGUGUGCAUCUUGUUUAAAGACGGUUCACUGGCAUUAGUAU-----UUGAUCUU ...((((((((.(((((.....((((........)))).(((.((((((((..((((((.(((((....)).))).))))))..)))))))).))).)).))).)))-----)))))... ( -28.50) >DroEre_CAF1 20401 108 - 1 AAAAUUAGAUAGUGAGCGGAAAUGGUAUU--CGAACCGAAGUAAAUCGUCCAAAAACAACAUGGC-----UGCAUCUUGUUUAAGGACGGUUCACUCGCCAUAUUAU-----ACGAUCUU ...........(((.(((....((((...--...)))).(((.((((((((..((((((.(((..-----..))).))))))..)))))))).))))))))).....-----........ ( -27.60) >DroYak_CAF1 15891 113 - 1 AAAAUUAGAUAGUGAGUGGAAAUGGUAUA--AGAACCGGAGUAAAUCGUCCAAAAACAACAUGGC-----UGCAUCUUGUUUAAAGACGGUUCACUCGCAAUAAUAUAUAUUAUGAUCAU .......(((.(((((((((..((((...--...)))).......(((((...((((((.(((..-----..))).))))))...)))))))))))))).(((((....))))).))).. ( -27.60) >consensus AAAAUUAGAUAGUGAGUGGAAAUGGUAUA__AGAACCGGAGUAAAUCGUCCAAAAACAACAUGGC_____UGCAUCUUGUUUAAAGACGGUUCACUCGCAAUAAUAU_____AUGAUCUU ...........(((((((((..((((........)))).......((((((..((((((.(((.........))).))))))..)))))))))))))))..................... (-23.35 = -24.13 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:24 2006