
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,209,382 – 11,209,518 |
| Length | 136 |
| Max. P | 0.958607 |

| Location | 11,209,382 – 11,209,501 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -24.19 |
| Energy contribution | -24.80 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
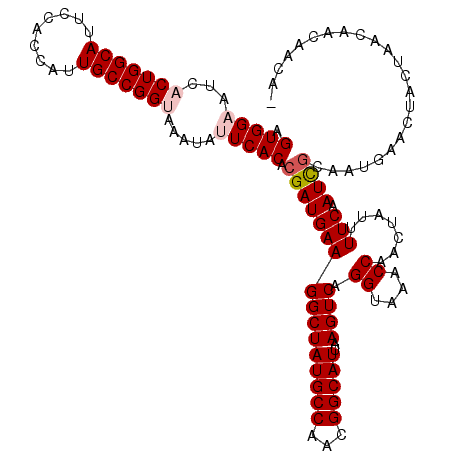
>3R_DroMel_CAF1 11209382 119 + 27905053 AGUGGAAUCACUGGCAUUCCACCCCUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCACCUAUUUUCAAUCGCAAUGAACUACGAACAACAACA- .((((....(((((((.........))))))).....((((..((((((((((((((((...)))))..))))((((.....))))...))).))))...))))))))...........- ( -30.40) >DroPse_CAF1 16202 117 + 1 AGUGGCAUCACUGGCAUCCCACCAUUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUCGCAAUGCAU--CUAACUACAACA- (((.((((.(((((((.........)))))))...........((((((((((((((((...)))))..)))).((....)).......))).))))..))))..--...)))......- ( -27.80) >DroGri_CAF1 12220 120 + 1 AGUGGAAUAUCUGGCAUUCCACCAUUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAAUUAUUUUCAAUUGCAAUGAACUACAAGCAACAAAAA .(((((((((((((((.........))))))...)))))).)))..(((((((((((((...)))))..)))).((....)).......)))).((((............))))...... ( -29.10) >DroWil_CAF1 14831 116 + 1 AGUGGAAUCACUGGCAUUCCACCAUUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUUGCAAUGAACUA---ACAACAAGA- .((((((((....).)))))))((((((((((........)).)).(((((((((((((...)))))..)))).((....)).......))))...)))))).....---.........- ( -26.60) >DroAna_CAF1 11715 119 + 1 AGUGGAAUCUCUGGCAUUCCACCUCUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCGACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUCACAAUGAACUAACAACAGCAACA- .(((((((..((((((.........)))))).....)))).)))(((((((((((((((...)))))..)))).((....)).......))).))).......................- ( -25.50) >DroPer_CAF1 16776 117 + 1 AGUGGCAUCACUGGCAUCCCACCAUUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUCGCAAUGCAU--CUAACUACAACA- (((.((((.(((((((.........)))))))...........((((((((((((((((...)))))..)))).((....)).......))).))))..))))..--...)))......- ( -27.80) >consensus AGUGGAAUCACUGGCAUUCCACCAUUGCCGGUAAAUAUUCACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUCGCAAUGAACUACUAACAACAACA_ .(((((...(((((((.........))))))).....))))).((((((((((((((((...)))))..)))).((....)).......))).))))....................... (-24.19 = -24.80 + 0.61)

| Location | 11,209,422 – 11,209,518 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.52 |
| Mean single sequence MFE | -15.27 |
| Consensus MFE | -12.22 |
| Energy contribution | -12.17 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11209422 96 + 27905053 ACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCACCUAUUUUCAAUCGCAAUGAACUACGAACAACAACA--GCAACC--ACA----GCAA-UUAU ...((((((((((((((((...)))))..))))((((.....))))...))).))))......................--((....--...----))..-.... ( -16.40) >DroVir_CAF1 10998 101 + 1 ACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUUGCAAUGAACUACAAGCAACAAAAACGAAACCCAACAA---ACAA-UUAU ...((.(((((((((((((...)))))..)))).((....)).......)))).((((............))))......))...........---....-.... ( -14.30) >DroGri_CAF1 12260 101 + 1 ACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAAUUAUUUUCAAUUGCAAUGAACUACAAGCAACAAAAAAGCAUCAAAAAAA---AAAA-AUAA ....((((..(((((((((...)))))..)))).((....))............((((............))))........)))).......---....-.... ( -15.30) >DroWil_CAF1 14871 96 + 1 ACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUUGCAAUGAACUA---ACAACAAGA--GCAACU--GCA-ACUGCAA-CUCA ......(((((((((((((...)))))..)))).((....)).......)))).(((((.((.....---.))......--((....--)).-..)))))-.... ( -16.30) >DroMoj_CAF1 11139 95 + 1 ACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUUGCAAUGAACUACAAGCAACAAAU--GAAA---AAUAU---ACUA-U-AU ..........(((((((((...)))))..)))).((....)).....((((((.((((............))))....)--))))---)....---....-.-.. ( -14.20) >DroAna_CAF1 11755 96 + 1 ACACGAUGAAGGCUAUGCCGACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUCACAAUGAACUAACAACAGCAACA--GCAACC--ACA----GCCAAAUA- ..........(((((((((...))))).......(((............((((.......)))).........((....--)).)))--..)----))).....- ( -15.10) >consensus ACACGAUGAAGGCUAUGCCAACGGCAUUAAGUCAGGUAAACCAACUAUUUUCAAUUGCAAUGAACUACAAACAACAAAA__GCAACC__ACA____ACAA_UUAU ...((((((((((((((((...)))))..)))).((....)).......))).))))................................................ (-12.22 = -12.17 + -0.05)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:17 2006