| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,531,730 – 1,531,823 |
| Length | 93 |
| Max. P | 0.991862 |

| Location | 1,531,730 – 1,531,823 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.74 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -11.36 |
| Energy contribution | -12.04 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
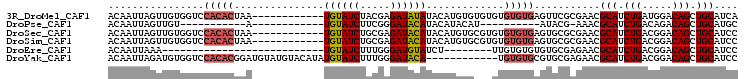
>3R_DroMel_CAF1 1531730 93 + 27905053 ACAAUUAGUUGUGGUCCACACUAA------------UGUAUCUACGAGAUAUAUACAUGUGUGUGUGUGUGAGUUCGCGAACGCAUCUGAUGGACAGCUGCAUCA .......((.((.(((((.....(------------((((((.....))))))).((.((((((...((((....)))).)))))).)).))))).)).)).... ( -26.80) >DroPse_CAF1 32366 71 + 1 ACAAUUAGUUGU-----------A------------UGUAUCUUCGGGAUACAUACAUACAU----------AUACG-AAACGCAUCUGACAGACAGCUGCAUGC .......(((((-----------(------------((((((.....)))))))))).))..----------.....-....(((.(((.....))).))).... ( -19.30) >DroSec_CAF1 33311 93 + 1 ACAAUUAGUUGUGGUCCACACUAA------------UGUAUCUGCGAGAUACAUACAUGUGCGUGUGUGUGAGUGCGCGAACGCAUCUGACGGACAGCUGCAUCC .......((.((.((((......(------------((((((.....))))))).((.((((((.(((((....))))).)))))).))..)))).)).)).... ( -31.50) >DroSim_CAF1 31573 93 + 1 ACAAUUAGUUGUGGUCCACACUAA------------UGUAUCUGCGAGAUACAUACAUGUGCGUGUGUGUGAGUGCGCGAACGCAUCUGACGGACAGCUGCAUCC .......((.((.((((......(------------((((((.....))))))).((.((((((.(((((....))))).)))))).))..)))).)).)).... ( -31.50) >DroEre_CAF1 32782 70 + 1 ACAAUUAAA---------------------------UGUAUCUUUGGGAUGUAUCU--------UUGUGUGUGUGCGAGAACGCAUCUGACGGACAGCUGCAUCC .........---------------------------..........(((((((...--------((((.((((((((....)))))...))).)))).))))))) ( -17.80) >DroYak_CAF1 29318 93 + 1 ACAAUUAGAUGUGGUCCACACGGAUGUAUGUACAUAUGUAUCUUUGGGAUACA------------UGUGUGCGUGCGAGAACGCAUCUGACGGACAGCUGCAUCC .(..(((((((((((((....))))(((((((((((((((((.....))))))------------))))))))))).....)))))))))..)............ ( -40.10) >consensus ACAAUUAGUUGUGGUCCACACUAA____________UGUAUCUUCGAGAUACAUACAUGUG___GUGUGUGAGUGCGCGAACGCAUCUGACGGACAGCUGCAUCC ................(((((...............((((((.....)))))).............)))))...........(((.(((.....))).))).... (-11.36 = -12.04 + 0.68)
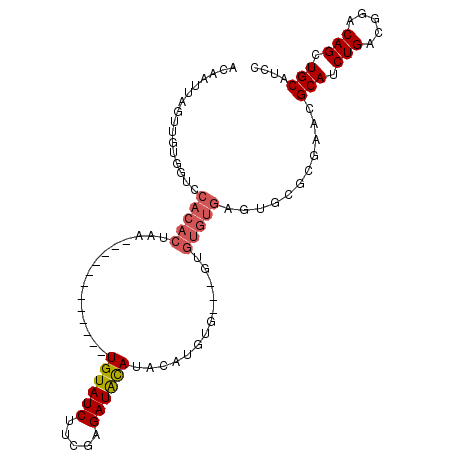
| Location | 1,531,730 – 1,531,823 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 69.74 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -13.23 |
| Energy contribution | -14.99 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1531730 93 - 27905053 UGAUGCAGCUGUCCAUCAGAUGCGUUCGCGAACUCACACACACACACAUGUAUAUAUCUCGUAGAUACA------------UUAGUGUGGACCACAACUAAUUGU .((((((.(((.....))).))))))..............(((((..((((((.(((...))).)))))------------)..)))))................ ( -18.80) >DroPse_CAF1 32366 71 - 1 GCAUGCAGCUGUCUGUCAGAUGCGUUU-CGUAU----------AUGUAUGUAUGUAUCCCGAAGAUACA------------U-----------ACAACUAAUUGU ..(((((.(((.....))).)))))..-.....----------.....((((((((((.....))))))------------)-----------)))......... ( -19.00) >DroSec_CAF1 33311 93 - 1 GGAUGCAGCUGUCCGUCAGAUGCGUUCGCGCACUCACACACACGCACAUGUAUGUAUCUCGCAGAUACA------------UUAGUGUGGACCACAACUAAUUGU ((((......))))(((.((((((....)))).)).......(((((....(((((((.....))))))------------)..))))))))............. ( -26.30) >DroSim_CAF1 31573 93 - 1 GGAUGCAGCUGUCCGUCAGAUGCGUUCGCGCACUCACACACACGCACAUGUAUGUAUCUCGCAGAUACA------------UUAGUGUGGACCACAACUAAUUGU ((((......))))(((.((((((....)))).)).......(((((....(((((((.....))))))------------)..))))))))............. ( -26.30) >DroEre_CAF1 32782 70 - 1 GGAUGCAGCUGUCCGUCAGAUGCGUUCUCGCACACACACAA--------AGAUACAUCCCAAAGAUACA---------------------------UUUAAUUGU (((((((.(((.....))).)))))))..........((((--------((((..(((.....)))..)---------------------------)))..)))) ( -11.60) >DroYak_CAF1 29318 93 - 1 GGAUGCAGCUGUCCGUCAGAUGCGUUCUCGCACGCACACA------------UGUAUCCCAAAGAUACAUAUGUACAUACAUCCGUGUGGACCACAUCUAAUUGU ((((......))))...(((((.(.((.((((((.....(------------((((((.....)))))))((((....)))).)))))))).).)))))...... ( -26.40) >consensus GGAUGCAGCUGUCCGUCAGAUGCGUUCGCGCACUCACACAC___CACAUGUAUGUAUCCCGAAGAUACA____________UUAGUGUGGACCACAACUAAUUGU (((((((.(((.....))).))))))).......(((((.............((((((.....))))))...............)))))................ (-13.23 = -14.99 + 1.76)

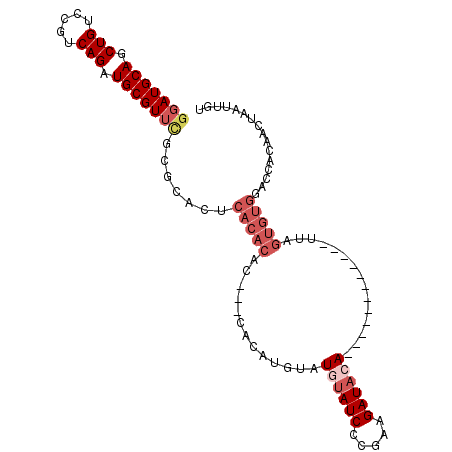
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:31 2006