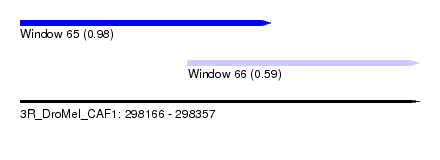
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 298,166 – 298,357 |
| Length | 191 |
| Max. P | 0.978834 |

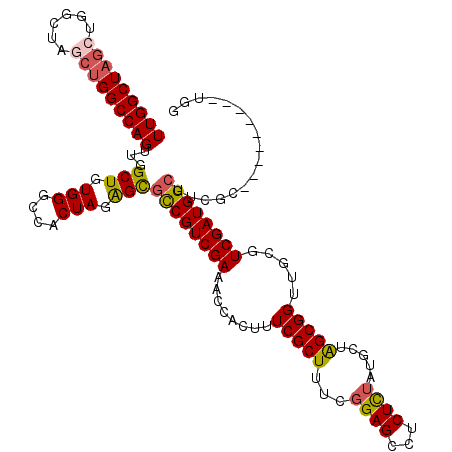
| Location | 298,166 – 298,286 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -41.64 |
| Consensus MFE | -38.78 |
| Energy contribution | -39.26 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 298166 120 + 27905053 UUGGUUUUCUGAUGUAACAGCGGUUGACGCGACAACUCGUUGACCAGUUGGCAAUUUCCAUUUCCAUCCAUGGCUACGAUUUGGCUAGGUGGCUAGCUGGCCAGUUGGCUGUGGGCCACU .((((.......(((..(((((((..(((.(....).)))..))).)))))))..............(((((((((....((((((((........)))))))).))))))))))))).. ( -40.60) >DroSec_CAF1 33614 120 + 1 UUGGCUUUCUGAUGUAACAGCGGUUGACGCGACAGCUCGUUGACCAGUUGGCAAUUUCCAUUUCCAUCCAUGGCUGCGAUUUGGCUAGCUAGCUAGCUGGCCAGUUGGCUGUGGGCCACU .((((.......(((..(((((((..(((.(....).)))..))).)))))))..............(((((((((....((((((((((....)))))))))).))))))))))))).. ( -46.20) >DroSim_CAF1 35301 120 + 1 UUGGCUUUCUGAUGUAACAGCGGUUGACGCGUCAGCUCGUUGACCAGUUGGCAAUUUCCAUUUCCAUCCAUGGCUGCGAUUUGGCUAGCUGGCUAGCUGGCCAGUUGGCUGUGGGCCACU .((((.......(((..(((((((..(((.(....).)))..))).)))))))..............(((((((((....((((((((((....)))))))))).))))))))))))).. ( -46.80) >DroEre_CAF1 36216 116 + 1 UUGGCUUUCUGAUGUAACAGCGGUUGACGCGACAGCCCGUUGACCAGUUGGCAAUUUCCAUUUCCAUUCAUGGCUGCGAUUUGGCUAGCUG----GCUGGCCAGUUGGCUGUGGGCCACU .((((((............(((.....))).((((((..(((.(((((..((.....(((..((((........)).))..)))...))..----))))).)))..)))))))))))).. ( -43.70) >DroYak_CAF1 37460 111 + 1 UU-UCUUUCUGAUGUAACAGCGGUUGACGCGACUACUUGUUGACCAGUUGGCAAUUUCCAUUUUCAUUCAUGGCUGCGAUUUGGCUAGCU--------GGCCAGUUGGCUGUGGGCCACU ..-..............(((((((..(((........)))..))).))))(((....((((........)))).)))....(((((.((.--------((((....)))))).))))).. ( -30.90) >consensus UUGGCUUUCUGAUGUAACAGCGGUUGACGCGACAGCUCGUUGACCAGUUGGCAAUUUCCAUUUCCAUCCAUGGCUGCGAUUUGGCUAGCUGGCUAGCUGGCCAGUUGGCUGUGGGCCACU .((((.......(((..(((((((..(((.(....).)))..))).)))))))..............(((((((((....(((((((((......))))))))).))))))))))))).. (-38.78 = -39.26 + 0.48)

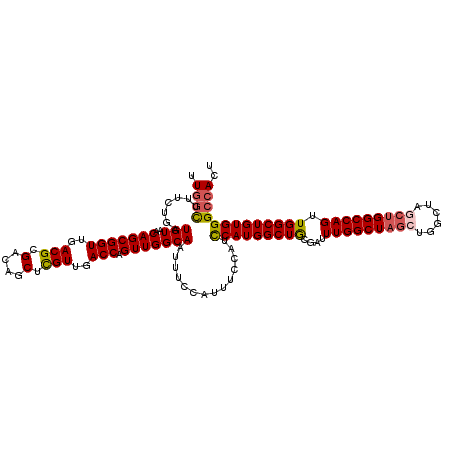
| Location | 298,246 – 298,357 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -45.04 |
| Consensus MFE | -36.36 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 298246 111 + 27905053 UUGGCUAGGUGGCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGCGUCGUCGAAACCACUUUCGCUUUCGGAGCCUCUCUAUGCUAGCGGUUUCGUCGAUGGCUUCGC---------UGG (..((..(((((((.((.((((....)))))).))))))).((((((((.(((((((.((...((....((((...))))..)).)).))))))).)))).)))).))---------..) ( -46.80) >DroSec_CAF1 33694 111 + 1 UUGGCUAGCUAGCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGCGCCGUCGAAACCACUUUCGCUUUCGGAGCCUCUCUAUGCUGGCGGUUGCGUCGAUGGCUUCGC---------UGG ((((((((((....)))))))))).((((.....))))((((...((((((((((((...((((....))))(((..........)))))))...))))))))....)---------))) ( -45.40) >DroSim_CAF1 35381 111 + 1 UUGGCUAGCUGGCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGCGCCGUCGAAACCACUUUCGCUUUCGGAGCCUCUCUAUGCUGGCGGUUGCGUCGAUGGCUUCGC---------UGG ..((((((((((((....))))))))))))((((((((...((((((((((((((....))))((....((((...))))..)).))))).))).))..)))).))))---------... ( -46.40) >DroEre_CAF1 36296 114 + 1 UUGGCUAGCUG----GCUGGCCAGUUGGCUGUGGGCCACUAGAGCGCCGUCGAAACCACUUUCGCUUUCGGAGCCUCUUUG--UUAGCGGUUGCGUCGAUGGCUUCGCUGGCGUCGCUGG ..(((((((((----(....))))))))))((((((((...(((.((((((((...((...(((((..(((((...)))))--..))))).))..)))))))))))..)))).))))... ( -51.70) >DroYak_CAF1 37539 106 + 1 UUGGCUAGCU--------GGCCAGUUGGCUGUGGGCCACUAGGGUACCGUCGAAAUCACUUUCGCUUUCUGAGCCUCUCUAUUUUAGCGGUUGCGUCGAUGGCUUCGC------CGCUAU .(((((.((.--------((((....)))))).))))).((((((.(((((((...((...(((((....(((...)))......))))).))..)))))))....))------).))). ( -34.90) >consensus UUGGCUAGCUGGCUAGCUGGCCAGUUGGCUGUGGGCCACUAGAGCGCCGUCGAAACCACUUUCGCUUUCGGAGCCUCUCUAUGCUAGCGGUUGCGUCGAUGGCUUCGC_________UGG (((((((((......)))))))))...(((.(((....))).)))((((((((........(((((...((((...)))).....))))).....))))))))................. (-36.36 = -36.68 + 0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:07 2006