
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,198,673 – 11,198,774 |
| Length | 101 |
| Max. P | 0.814251 |

| Location | 11,198,673 – 11,198,774 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -14.87 |
| Energy contribution | -14.98 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
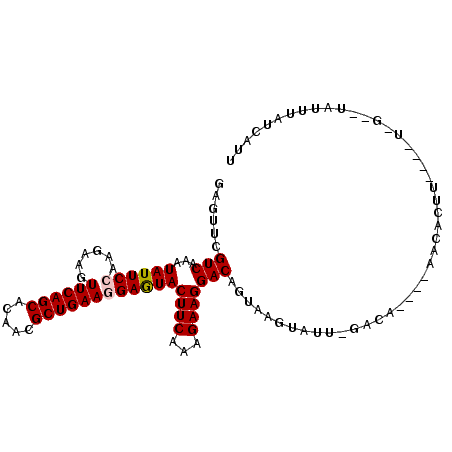
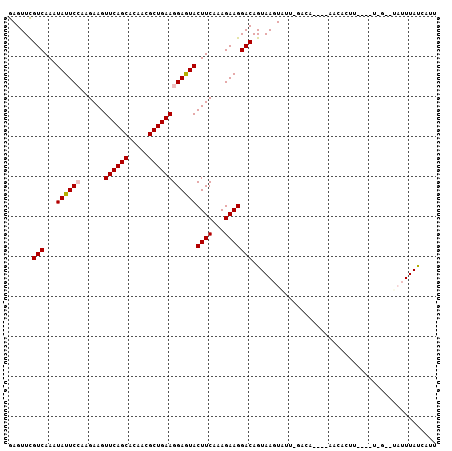
>3R_DroMel_CAF1 11198673 101 + 27905053 GAGUUCGUCAAAUAUUCCAAGAAGUUCAGCACAACGCUGAAGGAGUACUUCAAAGAAGGACAGUAAGUAUU-AAGA----AUCACCUGCAGUGA-UUAUUUACUAUA ......(((...((((((......((((((.....))))))))))))((((...)))))))((((((((..-....----(((((.....))))-)))))))))... ( -24.50) >DroVir_CAF1 770 86 + 1 GAGUUCGUCAAAUAUUCAAAGAAGUUCAGCACAACGCUGAAGGAAUACUUCAAAGAAGGACAGUAAGUUGG---C-------UCAUU------G-----UUAUCGUU ((((.........))))...((((((((((.....)))))......)))))...((..((((((.((....---)-------).)))------)-----)).))... ( -16.30) >DroPse_CAF1 5710 101 + 1 GAGUUUGUCAAAUAUUCCAAGAAGUUCAGCACAACGCUGAAGGAGUACUUCAAAGAAGGACAGUAAGUGUU-GUGACAAAGACACU-----UUGUCUAUUUAUCGUU .....((((...((((((......((((((.....))))))))))))((((...))))))))(((((((..-..(((((((...))-----)))))))))))).... ( -24.30) >DroGri_CAF1 1274 92 + 1 GAGUUCGUCAAAUAUUCAAAGAAGUUCAGCACAACGCUGAAGGAAUACUUCAAAGAAGGACAGUAAGUUGUUGGCA----ACCAUUU------G-----UUAUCAUU ((((.........))))...((((((((((.....)))))......)))))...((..(((((...((((....))----))...))------)-----)).))... ( -17.70) >DroAna_CAF1 1058 102 + 1 GAGUUUGUCAAAUAUUCCAAGAAGUUCAGCACAACGCUGAAGGAGUACUUCAAAGAAGGACAGUAAGUACAGAACA----UUAAGCUCCGGCCAUGUAUUUAC-AUU .....((((...((((((......((((((.....))))))))))))((((...))))))))(((((((((.....----....((....))..)))))))))-... ( -25.30) >DroPer_CAF1 5563 101 + 1 GAGUUUGUCAAAUAUUCCAAGAAGUUCAGCACAACGCUGAAGGAGUACUUCAAAGAAGGACAGUAAGUGUU-GUGACAAAGACACU-----UUGUCUAUUUAUCGUU .....((((...((((((......((((((.....))))))))))))((((...))))))))(((((((..-..(((((((...))-----)))))))))))).... ( -24.30) >consensus GAGUUCGUCAAAUAUUCCAAGAAGUUCAGCACAACGCUGAAGGAGUACUUCAAAGAAGGACAGUAAGUAUU_GACA____AACACUU____U_G__UAUUUAUCAUU ......(((...((((((......((((((.....))))))))))))((((...))))))).............................................. (-14.87 = -14.98 + 0.11)

| Location | 11,198,673 – 11,198,774 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.48 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.12 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
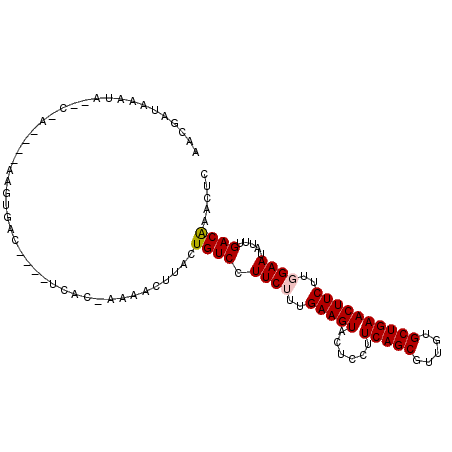
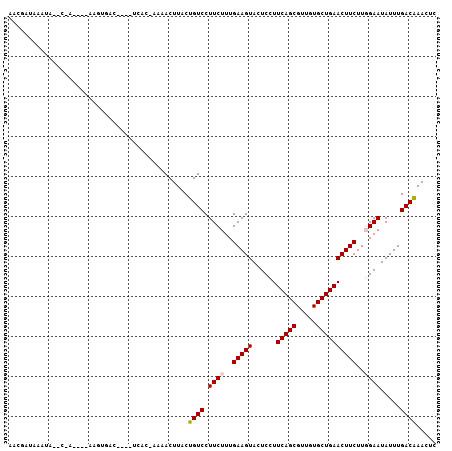
>3R_DroMel_CAF1 11198673 101 - 27905053 UAUAGUAAAUAA-UCACUGCAGGUGAU----UCUU-AAUACUUACUGUCCUUCUUUGAAGUACUCCUUCAGCGUUGUGCUGAACUUCUUGGAAUAUUUGACGAACUC ...((((((.((-((((.....)))))----).))-..))))...((((.((((..(((((......(((((.....))))))))))..)))).....))))..... ( -20.20) >DroVir_CAF1 770 86 - 1 AACGAUAA-----C------AAUGA-------G---CCAACUUACUGUCCUUCUUUGAAGUAUUCCUUCAGCGUUGUGCUGAACUUCUUUGAAUAUUUGACGAACUC ..((...(-----(------(.(((-------(---....)))).)))..(((...(((((......(((((.....))))))))))...))).......))..... ( -14.00) >DroPse_CAF1 5710 101 - 1 AACGAUAAAUAGACAA-----AGUGUCUUUGUCAC-AACACUUACUGUCCUUCUUUGAAGUACUCCUUCAGCGUUGUGCUGAACUUCUUGGAAUAUUUGACAAACUC ...(.(((((((((((-----(((((.........-.))))))..)))).((((..(((((......(((((.....))))))))))..)))))))))).)...... ( -22.20) >DroGri_CAF1 1274 92 - 1 AAUGAUAA-----C------AAAUGGU----UGCCAACAACUUACUGUCCUUCUUUGAAGUAUUCCUUCAGCGUUGUGCUGAACUUCUUUGAAUAUUUGACGAACUC ........-----(------(((((((----((....)))).........(((...(((((......(((((.....))))))))))...)))))))))........ ( -16.10) >DroAna_CAF1 1058 102 - 1 AAU-GUAAAUACAUGGCCGGAGCUUAA----UGUUCUGUACUUACUGUCCUUCUUUGAAGUACUCCUUCAGCGUUGUGCUGAACUUCUUGGAAUAUUUGACAAACUC .((-((....))))(..((((((....----.))))))..)....((((.((((..(((((......(((((.....))))))))))..)))).....))))..... ( -23.40) >DroPer_CAF1 5563 101 - 1 AACGAUAAAUAGACAA-----AGUGUCUUUGUCAC-AACACUUACUGUCCUUCUUUGAAGUACUCCUUCAGCGUUGUGCUGAACUUCUUGGAAUAUUUGACAAACUC ...(.(((((((((((-----(((((.........-.))))))..)))).((((..(((((......(((((.....))))))))))..)))))))))).)...... ( -22.20) >consensus AACGAUAAAUA__C_A____AAGUGAC____UCAC_AAAACUUACUGUCCUUCUUUGAAGUACUCCUUCAGCGUUGUGCUGAACUUCUUGGAAUAUUUGACAAACUC .............................................((((.((((..(((((......(((((.....))))))))))..)))).....))))..... (-13.03 = -13.12 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:05 2006