
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,189,820 – 11,190,002 |
| Length | 182 |
| Max. P | 0.977987 |

| Location | 11,189,820 – 11,189,911 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 66.35 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11189820 91 - 27905053 ---------------------AUACUCAAAUACAUACAUAUUCAGAGGUGCCAUGAGCAAGGCUCCACCAACCUAUCCAUAGAUCCGUGGGAUUGGAGCAUCCGUCUAUGGG ---------------------...(((.((((......))))..)))...(((((..(...((((((((.((..(((....)))..)).))..))))))....)..))))). ( -21.40) >DroSim_CAF1 2313 112 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNAGGUGCCAUGAGCAAGGCUCCACCAACCUAUCCAUAGAUCCUUGGGAUUGGAGCAGUAGUCUAUGGG ..................................................(((((..((..((((((((((...(((....)))..))))...))))))..).)..))))). ( -19.80) >DroYak_CAF1 2346 88 - 1 ---------------------AUACUCAAAUACAUACAUAUUCAGAGGUGCCAUGAGCAAGGCUCC---AACCUAUCCAUAGAUCCGUGGGAUUGGAGCAUCCGUCUCUGGG ---------------------....................((((((((((.....)))..(((((---(((((((..........))))).))))))).....))))))). ( -25.00) >consensus _____________________AUACUCAAAUACAUACAUAUUCAGAGGUGCCAUGAGCAAGGCUCCACCAACCUAUCCAUAGAUCCGUGGGAUUGGAGCAUCCGUCUAUGGG .............................................((((.....((((...)))).....))))..((((((((..(((.........)))..)))))))). (-18.28 = -18.17 + -0.11)

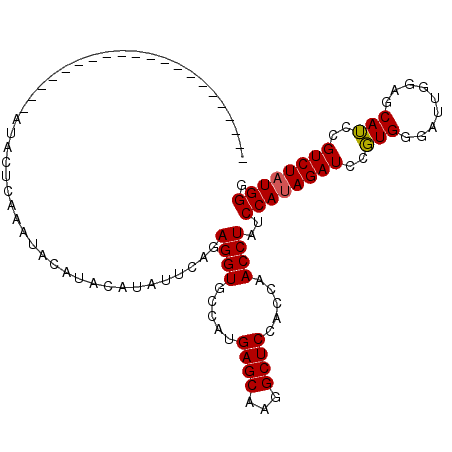
| Location | 11,189,911 – 11,190,002 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.35 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -19.53 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11189911 91 + 27905053 -----------------------------GUAGUUACUGCAGCCUUAUCUGCGAGGCAGGGACACCGCCCAAGCCCCAGCCGACUGCAAUUAAGUCGCAUGGCACAUUCGUGAUACUGUC -----------------------------(((.((((.((.(((((......))))).(((......)))..)).(((..(((((.......)))))..))).......))))))).... ( -25.30) >DroSim_CAF1 2625 119 + 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNGGGGCAGGGACACUGCCCAAGCCCCAGCCGACUGCAAU-AAGUCGCAUGGCACAUUCGUGAUACUGUC .....................................................((((((.....)))))).....(((..(((((.....-.)))))..)))(((....)))........ ( -23.80) >DroYak_CAF1 2434 91 + 1 -----------------------------GUAGUUGCUGCAGCCUUAUCUGUGGGGUAGGGACACCGCCCAAGCCCCAACCGACUGCAAUUUAGUCGCAUGGCACAUUCGUGAUACUGUC -----------------------------(((((((..((((......))))(((((.(((......)))..)))))...)))))))....((((..(((((.....)))))..)))).. ( -29.10) >consensus _____________________________GUAGUU_CUGCAGCCUUAUCUG_GGGGCAGGGACACCGCCCAAGCCCCAGCCGACUGCAAUUAAGUCGCAUGGCACAUUCGUGAUACUGUC ......................................((((......))))(((((.(((......)))..)))))...(((((.......)))))(((((.....)))))........ (-19.53 = -19.87 + 0.34)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:01 2006