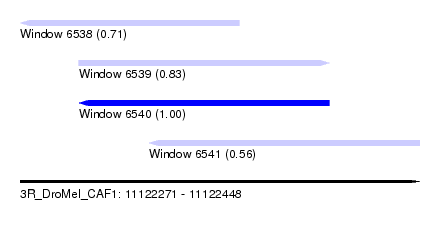
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,122,271 – 11,122,448 |
| Length | 177 |
| Max. P | 0.996389 |

| Location | 11,122,271 – 11,122,368 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -25.97 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11122271 97 - 27905053 GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUAU------A--AUAUAGUGUGGAUGGUA----GAGUAUAUAGACCC--CAU ..((((((.(((...........))).))))))((.(((.....(((((((.(((((((((.------.--.))))))))...).)))----))))....))).))--... ( -28.50) >DroSec_CAF1 32757 94 - 1 GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUAU------G--AUAUAGUGUGGAUU-------CAAUAUAUAGACCC--CAU ..((((((.(((...........))).))))))((((.....((((((.....((((((((.------.--.)))))))))))))-------)........)))).--... ( -24.92) >DroSim_CAF1 31903 93 - 1 GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAU-U------G--AUAUAGUGUGGAUU-------CAAUAUAUAGACCC--CAU ..((((((.(((...........))).))))))((((.....((((((.....(((((((-.------.--..))))))))))))-------)........)))).--... ( -25.02) >DroWil_CAF1 36745 87 - 1 GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUACGAGCACUAUAUGUA---------U-----ACAU--------AUAUAUGCAGAUCA--CAA ..((((((.(((...........))).))))))(((((....)))))......(((.(((((((.---------.-----..))--------))))))))......--... ( -25.40) >DroYak_CAF1 33766 109 - 1 GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUAUAUAUCGUAUAUAUAGUAGAGAUGGUAGAGUACAUAUAUAGACCC--CAU ..((((((.(((...........))).))))))((((.......(((((((.(.(((((((((((...)))))))))).)...).))))))).........)))).--... ( -28.99) >DroAna_CAF1 33014 85 - 1 GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUCUUGCGAGCACUAUAU-------------A---------GUU----GAAUAUAUAUAUCGGAUAU ..((((((.(((...........))).)))))).((((.((((...)))).))))..(((((-------------(---------...----...)))))).......... ( -23.00) >consensus GAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUAU_________AUAUAGUGUGGAU________AAAUAUAUAGACCC__CAU ..((((((.(((...........))).)))))).((((.((((...)))).))))........................................................ (-18.24 = -18.35 + 0.11)

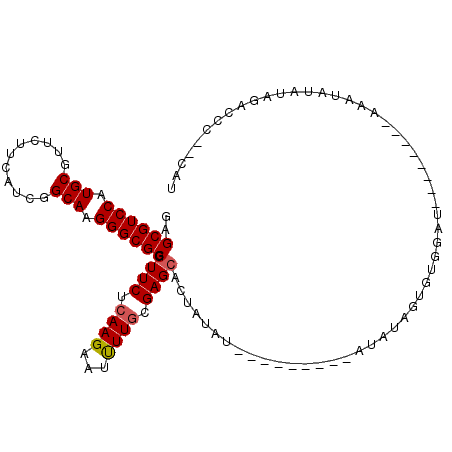
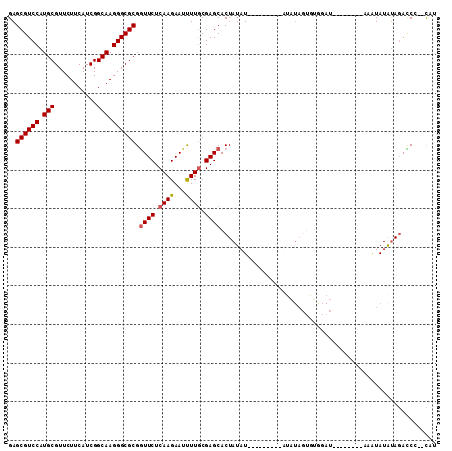
| Location | 11,122,297 – 11,122,408 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -31.01 |
| Energy contribution | -31.32 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11122297 111 + 27905053 CACUAUAU--U------AUAUAGUGCUCGCAAAAUUCUUGAGAACCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCUCUCGAGAAUCAGCUGAAGGAAGCGCGUUUCCUUGCUGAGGA (((((((.--.------.))))))).........(((((((((...(((.((.(((...........))).)).))))))))))))(((((..(((((((....))))))))))))... ( -36.80) >DroVir_CAF1 37648 107 + 1 ------------UGUAUAUAUAGUGCUCGCAAGAUUCUUGAGAAUCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCACUUGAGAAUCAGCUGAAGGAAGCGCGUUUCCUUGCUGAGGA ------------.............(((....((((((..((....(((.((.(((...........))).)).))).))..))))))(((..(((((((....))))))))))))).. ( -31.80) >DroGri_CAF1 36955 107 + 1 ------------AAUAUAUAUAGUGCUCGCAAGAUUCUUGAGAAUCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCACUUGAGAAUCAGCUGAAGGAAGCGCGUUUCCUUGCUGAGGA ------------.............(((....((((((..((....(((.((.(((...........))).)).))).))..))))))(((..(((((((....))))))))))))).. ( -31.80) >DroWil_CAF1 36766 106 + 1 ----A---------UACAUAUAGUGCUCGUAAGAUUCUUGAGAAUCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCUCUUGAGAAUCAAUUGAAGGAAGCACGUUUCCUUGCUGAGGA ----.---------...........(((....((((((..(((...(((.((.(((...........))).)).))))))..)))))).....(((((((....)))))))...))).. ( -31.70) >DroMoj_CAF1 38099 107 + 1 ------------ACUAUAUACAGUGCACGCAAGAUUCUUGAGAAUCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCUCUUGAGAAUCAGCUGAAGGAAGCGCGUUUCCUUGCUGAGGA ------------........((((.((.((..((((((..(((...(((.((.(((...........))).)).))))))..)))))).))))(((((((....))))))))))).... ( -33.50) >DroPer_CAF1 39060 119 + 1 UAUUCAAUAAAAAAUAUAUAUAGUGCUCGCAAGAUUCUUGAGAAUCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCUCUCGAGAAUCAGCUGAAGGAAGCGCGUUUCCUUGCUGAGGA .........................(((....(((((((((((...(((.((.(((...........))).)).))))))))))))))(((..(((((((....))))))))))))).. ( -36.00) >consensus ____________A_UAUAUAUAGUGCUCGCAAGAUUCUUGAGAAUCGCGCCCUUGCCGAUGAAGAACGCAUGGACGCUCUUGAGAAUCAGCUGAAGGAAGCGCGUUUCCUUGCUGAGGA .........................(((((..(((((((((((...(((.((.(((...........))).)).)))))))))))))).....(((((((....))))))))).))).. (-31.01 = -31.32 + 0.31)

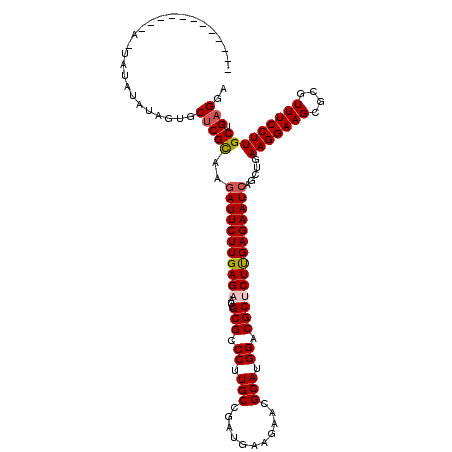
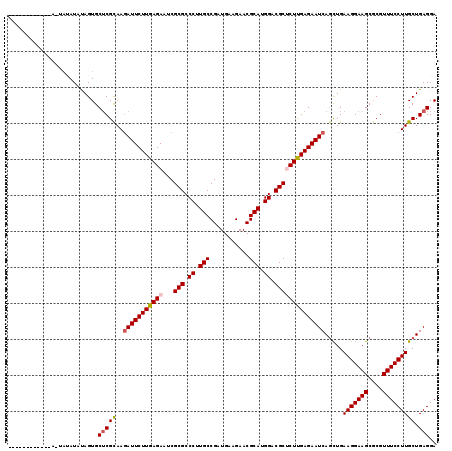
| Location | 11,122,297 – 11,122,408 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -32.36 |
| Energy contribution | -32.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11122297 111 - 27905053 UCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUCAAGAAUUUUGCGAGCACUAUAU------A--AUAUAGUG ..(((.(((((....(((((..((((..((((((....((((((((....)))))))).)))))))))))))))((((....))))))))))))(((((((.------.--.))))))) ( -39.10) >DroVir_CAF1 37648 107 - 1 UCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGUGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUAUAUACA------------ ...(((((((((((......))))))..)))))......(((((.((((.(((((((((....).))))))))(((((....))))).)).))))))).........------------ ( -33.70) >DroGri_CAF1 36955 107 - 1 UCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGUGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUAUAUAUU------------ ...(((((((((((......))))))..)))))......(((((.((((.(((((((((....).))))))))(((((....))))).)).))))))).........------------ ( -33.70) >DroWil_CAF1 36766 106 - 1 UCCUCAGCAAGGAAACGUGCUUCCUUCAAUUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUACGAGCACUAUAUGUA---------U---- ......(((.(....)((((((.........((((((..(((((((((.(((...........))).))))))...)))..))))))....))))))....))).---------.---- ( -30.42) >DroMoj_CAF1 38099 107 - 1 UCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGUGCACUGUAUAUAGU------------ ....(((...(....)((((..((((..((((((....((((((((....)))))))).))))))))))(((.(((((....))))).))))))).)))........------------ ( -35.30) >DroPer_CAF1 39060 119 - 1 UCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUAUAUAUUUUUUAUUGAAUA ..(((.(((((....)((((..((((..((((((....((((((((....)))))))).))))))))))))))(((((....))))))))))))......................... ( -37.00) >consensus UCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUCAAGAAUCUUGCGAGCACUAUAUAUA_U____________ ..(((.(((((....(((((..((((..((((((....((((((((....)))))))).)))))))))))))))((((....))))))))))))......................... (-32.36 = -32.42 + 0.06)


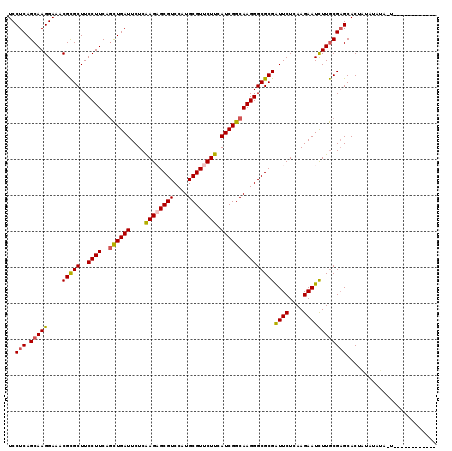
| Location | 11,122,328 – 11,122,448 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -31.08 |
| Energy contribution | -31.16 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11122328 120 - 27905053 AUUGGUUUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCGAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUC ..........((((((.((...........((((((.......(((((((((((......))))))..))))).....((((((((....))))))))....)))))))).))))))... ( -34.70) >DroVir_CAF1 37675 120 - 1 AUUGGUAUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGUGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUC .((((...(((((((........(((.((((((((...........))))))))))).........)))))))...)))).(((((((.(((...........))).)))))))...... ( -31.83) >DroGri_CAF1 36982 120 - 1 AUUGGUAUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGUGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUC .((((...(((((((........(((.((((((((...........))))))))))).........)))))))...)))).(((((((.(((...........))).)))))))...... ( -31.83) >DroWil_CAF1 36792 120 - 1 AUUGGUUUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGUGCUUCCUUCAAUUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUC ...(((.(......).)))..(((((..((((((((.........((((.(....).))))..........(((....((((((((....)))))))).))))))))))).))))).... ( -28.00) >DroMoj_CAF1 38126 120 - 1 AUUGGUAUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUC ...........((((((...............)).)))).((((.....))))..(((((..((((..((((((....((((((((....)))))))).)))))))))))))))...... ( -33.26) >DroAna_CAF1 33059 120 - 1 AUUGGUUUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGGUUCUC ..........((((((.((...........((((((.......(((((((((((......))))))..))))).....((((((((....))))))))....)))))))).))))))... ( -34.30) >consensus AUUGGUAUUUAGCUGCACCUCAUCGUAUUUCUUGUCAGCCUCCUCAGCAAGGAAACGCGCUUCCUUCAGCUGAUUCUCAAGAGCGUCCAUGCGUUCUUCAUCGGCAAGGGCGCGAUUCUC ...........((((((...............)).)))).((((.....))))..(((((..((((..((((((....((((((((....)))))))).)))))))))))))))...... (-31.08 = -31.16 + 0.08)

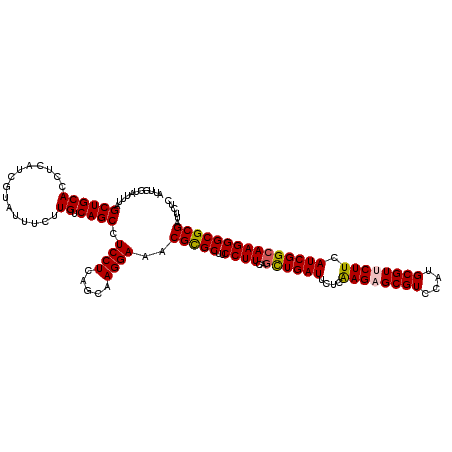
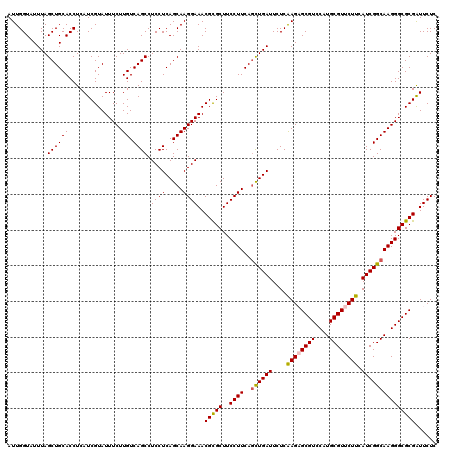
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:43 2006