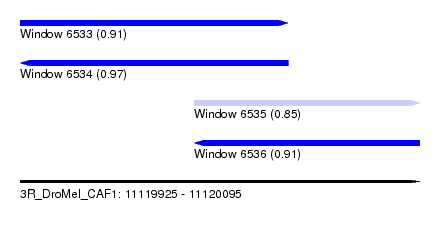
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,119,925 – 11,120,095 |
| Length | 170 |
| Max. P | 0.968824 |

| Location | 11,119,925 – 11,120,039 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -19.11 |
| Energy contribution | -21.25 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11119925 114 + 27905053 CUAUAUCCAUCUCUAUCUCCAUGCCCG-G--CACCAUAUCAGUC--AUUGAGCAUGGAUUUGGACAUGGACAUGG-CAUGGAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCCU (((..(((((..((((.((((((.(((-(--..((((.((((..--.))))..))))..)))).)))))).))))-.)))))...)))....((((..((((......))))...)))). ( -38.10) >DroPse_CAF1 36243 90 + 1 CUCUAUCUA------------UGUCCG-----ACCAUAUCAGUCACAUUGGACAUGGACAUGGACAUGGACAUGG------------GGACUG-GACUGGACUGCCUUGUCCAAUUGCUC (((((((((------------((((((-----.((((....(((......)))))))...))))))))))..)))------------))..((-(((.((....))..)))))....... ( -29.80) >DroSim_CAF1 29538 114 + 1 CUAUAUCCAUCUCUAUCUCCAUGCCCG-G--CACCAUAUCAGUC--AUUGGACAUGGAUUUGGACAUGGACACGG-CAUGGAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCCU ...........((((..((((((((.(-(--(.........)))--..((..((((........))))..)).))-))))))...))))...((((..((((......))))...)))). ( -37.00) >DroEre_CAF1 30854 115 + 1 CUAUAUCCAUCUCUAUCGCCAUGCCCG-G--CACCAUAUCAGUC--AUUGGACAUGGAUUUGGACAUGGACAUGGACAUGGAACUUAGAAGUGGCACCGGACUGCCUUGUCCAAUUGCUU (((..(((((.(((((..(((((.(((-(--..((((.((....--....)).))))..)))).)))))..))))).)))))...)))....((((..((((......))))...)))). ( -36.50) >DroYak_CAF1 31341 115 + 1 CUAUAUCCAUCUCUAUCGCCAUGCCUG-G--CACCAUAUCAGUC--AUUGGACAUGGAUUUGGACAUGGACAUGGACAUGGAACUUGGAACUGGCACCGGACUGCCUUGUCCAAUUGCUU ((((.(((((.(((((.((((....))-)--).(((.(((.(((--....)))...))).)))..))))).))))).))))...(((((...((((......))))...)))))...... ( -37.60) >DroAna_CAF1 30459 105 + 1 CUGUAUCUGUAUCUGUAUCUAUGCCCCGGCACACCAUAUCAGUC--AUUGGACAUGGUUUUGGACAUGGCACUGC-CAC------------CGGACCUGGACUGCCUUGUCCAAUUGCUU ..((((..(((....)))..))))...((((.(((((.((....--....)).))))).(((((((.((((...(-((.------------......)))..)))).))))))).)))). ( -27.90) >consensus CUAUAUCCAUCUCUAUCUCCAUGCCCG_G__CACCAUAUCAGUC__AUUGGACAUGGAUUUGGACAUGGACAUGG_CAUGGAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCUU ((((.(((((.((((...(((((.(((.....((.......)).....))).)))))...)))).))))).)))).................((((..((((......))))...)))). (-19.11 = -21.25 + 2.14)

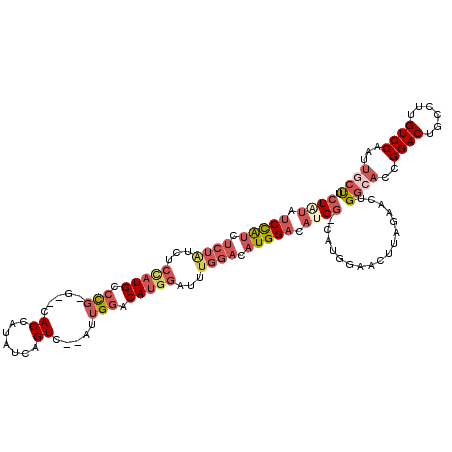

| Location | 11,119,925 – 11,120,039 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -23.35 |
| Energy contribution | -25.01 |
| Covariance contribution | 1.65 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11119925 114 - 27905053 AGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUG-CCAUGUCCAUGUCCAAAUCCAUGCUCAAU--GACUGAUAUGGUG--C-CGGGCAUGGAGAUAGAGAUGGAUAUAG .((((.((((((......))))))))))....(((...(((((.-(.((.(((((((((....(((((.(((..--...))))))))..--.-.))))))))).)).)..)))))..))) ( -43.60) >DroPse_CAF1 36243 90 - 1 GAGCAAUUGGACAAGGCAGUCCAGUC-CAGUCC------------CCAUGUCCAUGUCCAUGUCCAUGUCCAAUGUGACUGAUAUGGU-----CGGACA------------UAGAUAGAG .....(((((((......))))))).-..((((------------....(.(((((((...((((((.....))).))).))))))).-----))))).------------......... ( -27.30) >DroSim_CAF1 29538 114 - 1 AGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUG-CCGUGUCCAUGUCCAAAUCCAUGUCCAAU--GACUGAUAUGGUG--C-CGGGCAUGGAGAUAGAGAUGGAUAUAG .((((.((((((......))))))))))....(((...(((((.-(..(.(((((((((....(((((((....--....)))))))..--.-.))))))))).)..)..)))))..))) ( -46.10) >DroEre_CAF1 30854 115 - 1 AAGCAAUUGGACAAGGCAGUCCGGUGCCACUUCUAAGUUCCAUGUCCAUGUCCAUGUCCAAAUCCAUGUCCAAU--GACUGAUAUGGUG--C-CGGGCAUGGCGAUAGAGAUGGAUAUAG ..(((.((((((......))))))))).....(((...(((((.((.((..((((((((....(((((((....--....)))))))..--.-.))))))))..)).)).)))))..))) ( -41.10) >DroYak_CAF1 31341 115 - 1 AAGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCCAAGUUCCAUGUCCAUGUCCAUGUCCAAAUCCAUGUCCAAU--GACUGAUAUGGUG--C-CAGGCAUGGCGAUAGAGAUGGAUAUAG ......(((((...((((......))))...)))))..(((((.((.((..(((((((.....(((((((....--....)))))))..--.-..)))))))..)).)).)))))..... ( -37.60) >DroAna_CAF1 30459 105 - 1 AAGCAAUUGGACAAGGCAGUCCAGGUCCG------------GUG-GCAGUGCCAUGUCCAAAACCAUGUCCAAU--GACUGAUAUGGUGUGCCGGGGCAUAGAUACAGAUACAGAUACAG ......(((((((.((((.((((......------------.))-).).)))).))))))).((((((((....--....))))))))((((....)))).................... ( -33.80) >consensus AAGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUCCAUG_CCAUGUCCAUGUCCAAAUCCAUGUCCAAU__GACUGAUAUGGUG__C_CGGGCAUGGAGAUAGAGAUGGAUAUAG .....(((((((......))))))).............(((((........((((((((....(((((((..........))))))).......))))))))........)))))..... (-23.35 = -25.01 + 1.65)



| Location | 11,119,999 – 11,120,095 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -17.22 |
| Energy contribution | -17.18 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.80 |
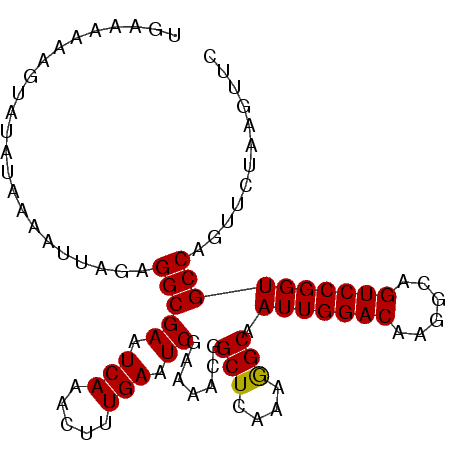
| SVM RNA-class probability | 0.853126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11119999 96 + 27905053 GAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCCUUUGAGCCGUUUACGAUUCAAAGUUUGAUUCGCCUCUAAUUUUAUAUACUUUUUUCA (((.(((((...(((...((((......))))......((((((..((....))..))))))........)))))))).))).............. ( -21.90) >DroSec_CAF1 30467 96 + 1 GAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCCUUUGAGCCGUUUUCGAUUCAAUGUUUGAUUCGCCUCUAAUUUUAUAUACUUUUUUCA (((...((....((((..((((......))))...))))...(((.((...((((........))))..)).)))............))...))). ( -19.60) >DroSim_CAF1 29612 96 + 1 GAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCCUUUGAGCCGUUUUCGAUUCAAAGUUUGAUUCGCCUCUAAUUUUAUAUACUUUUUUCA (((.(((((...(((...((((......))))......((((((..((....))..))))))........)))))))).))).............. ( -21.50) >DroEre_CAF1 30929 96 + 1 GAACUUAGAAGUGGCACCGGACUGCCUUGUCCAAUUGCUUUUCAGCCGUUUUCGAUUCAAAGUUUGAUUCGCCUUUAAUUUUAUAUACUUUUUUCA (((...(((((((((...((((......))))...((.....))))).....(((.(((.....))).)))...............))))))))). ( -16.70) >DroYak_CAF1 31416 96 + 1 GAACUUGGAACUGGCACCGGACUGCCUUGUCCAAUUGCUUUUCAGCCGUUUUCGAUUCAAAGUUUGAUUCGCCUUUAAUUUUAUAUACUUUUUUCA (((..(((....((((..((((......))))...))))......)))..)))((.(((.....))).)).......................... ( -17.40) >consensus GAACUUAGAACUGGCACCGGACUGCCUUGUCCAAUUGCCUUUGAGCCGUUUUCGAUUCAAAGUUUGAUUCGCCUCUAAUUUUAUAUACUUUUUUCA (((.(((((...(((...((((......))))......((((((..((....))..))))))........)))))))).))).............. (-17.22 = -17.18 + -0.04)



| Location | 11,119,999 – 11,120,095 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -22.16 |
| Consensus MFE | -18.12 |
| Energy contribution | -17.88 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11119999 96 - 27905053 UGAAAAAAGUAUAUAAAAUUAGAGGCGAAUCAAACUUUGAAUCGUAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUC ..................((((((((........((((((..((....))..))))))...(((((((......))))))))))...))))).... ( -24.90) >DroSec_CAF1 30467 96 - 1 UGAAAAAAGUAUAUAAAAUUAGAGGCGAAUCAAACAUUGAAUCGAAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUC ..................((((((.(..........((((..((....))..)))).((((.((((((......)))))))))).))))))).... ( -22.80) >DroSim_CAF1 29612 96 - 1 UGAAAAAAGUAUAUAAAAUUAGAGGCGAAUCAAACUUUGAAUCGAAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUC ..................((((((((........((((((..((....))..))))))...(((((((......))))))))))...))))).... ( -24.10) >DroEre_CAF1 30929 96 - 1 UGAAAAAAGUAUAUAAAAUUAAAGGCGAAUCAAACUUUGAAUCGAAAACGGCUGAAAAGCAAUUGGACAAGGCAGUCCGGUGCCACUUCUAAGUUC .......................(((((.(((.....))).)).......(((....))).(((((((......))))))))))............ ( -19.50) >DroYak_CAF1 31416 96 - 1 UGAAAAAAGUAUAUAAAAUUAAAGGCGAAUCAAACUUUGAAUCGAAAACGGCUGAAAAGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCCAAGUUC .......................(((((.(((.....))).)).......(((....))).(((((((......))))))))))............ ( -19.50) >consensus UGAAAAAAGUAUAUAAAAUUAGAGGCGAAUCAAACUUUGAAUCGAAAACGGCUCAAAGGCAAUUGGACAAGGCAGUCCGGUGCCAGUUCUAAGUUC .......................(((((.(((.....))).)).......(((....))).(((((((......))))))))))............ (-18.12 = -17.88 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:38 2006