
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,115,881 – 11,116,004 |
| Length | 123 |
| Max. P | 0.686028 |

| Location | 11,115,881 – 11,115,973 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.90 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -11.34 |
| Energy contribution | -11.63 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11115881 92 + 27905053 GCAAC---ACA-------GUUUGAAA---AGCCAAAAGGAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACAGUGCAGUGCCGCAGUGCCGCGGCCAAGCUCGA .....---..(-------(((((...---........((.....))(((((....)))))...((((......))))...(((((......)))))))))))... ( -27.30) >DroSec_CAF1 26314 83 + 1 GCAAC---ACA-------GUUUGAAA---AGCCAAAAGGAAAACCCGGCCAUUUAUGGUCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGA .....---..(-------(((((...---.(((....((...((..(((((....)))))...((((.....)---------))).)).))..)))))))))... ( -19.30) >DroSim_CAF1 24951 83 + 1 GCAAC---ACA-------GUUUGAAA---AGCCAAAAGGAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGA .....---..(-------(((((...---.(((....((...((..(((((....)))))...((((.....)---------))).)).))..)))))))))... ( -22.00) >DroEre_CAF1 26767 83 + 1 GCAAC---ACA-------GUUUGAAA---AGCCAAAAGGAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGA .....---..(-------(((((...---.(((....((...((..(((((....)))))...((((.....)---------))).)).))..)))))))))... ( -22.00) >DroYak_CAF1 27227 86 + 1 GCAACUGAGCA-------GUUUGAAA---AACCAAAAGGAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGA .....(((((.-------.((((...---...)))).((.....))((((.....((((.(..((((.....)---------)))..))))).))))..))))). ( -22.40) >DroPer_CAF1 31697 87 + 1 GCUA-----CAUGCUUCUACUACAAAUAUCACUAAAAGGAAAACAUUACCAUCUAUGGCCAAAGCAUAGCGCA----AA---UGCUGUA------CAAAGCCCGA ((((-----..(((((............((........))........(((....)))...)))))))))...----..---.(((...------...))).... ( -10.60) >consensus GCAAC___ACA_______GUUUGAAA___AGCCAAAAGGAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA_________UGCAGUGCCGCGGCCAAGCUCGA ((.............................((....)).......(((((....)))))...))..................((...((....))...)).... (-11.34 = -11.63 + 0.29)

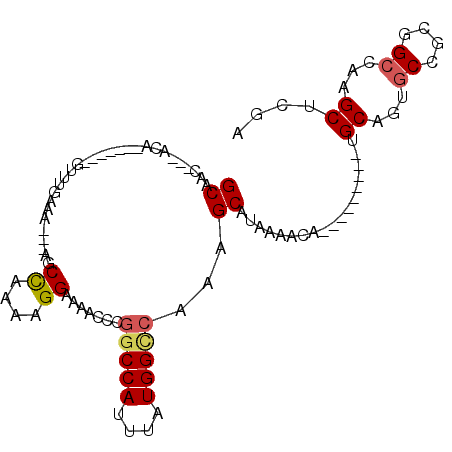
| Location | 11,115,906 – 11,116,004 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.22 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11115906 98 + 27905053 GAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACAGUGCAGUGCCGCAGUGCCGCGGCCAAGCUCGAUUUGCCAAAAUUGGCCAACAAAUUAAUUAUU ........(((((.((.((((....((((......)))).(.(((((......))))))...........)))).)).)))))............... ( -31.00) >DroPse_CAF1 31613 84 + 1 GAAAACAUUACCAUCUAUGGCCAAAGCAUAGCGCA----AA---UGCUGUA------CAAAGCCCGACUUGCCA-AUUUGGUGUACAAAUUAAUUAUU ........(((((....((((....(.((((((..----..---)))))).------).(((.....)))))))-...)))))............... ( -14.10) >DroSec_CAF1 26339 89 + 1 GAAAACCCGGCCAUUUAUGGUCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGAUUUGCCAAAAUUGGCCAACAAAUUAAUUAUU ........(((((.((.(((.((((((((.....)---------))).(.((....)))........))))))).)).)))))............... ( -22.30) >DroEre_CAF1 26792 89 + 1 GAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGAUUUUCCAAAAUUGGCCAACAAAUUAAUUAUU ........(((((....)))))...((((.....)---------)))........((((((....(......)....))))))............... ( -21.20) >DroYak_CAF1 27255 89 + 1 GAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA---------UGCAGUGCCGCGGCCAAGCUCGAUUUGCCAAAAUUGGCCAACAAAUUAAUUAUU ........(((((.((.((((....((((.....)---------))).(.((....)))...........)))).)).)))))............... ( -22.40) >DroPer_CAF1 31730 84 + 1 GAAAACAUUACCAUCUAUGGCCAAAGCAUAGCGCA----AA---UGCUGUA------CAAAGCCCGACUUGCCA-AUUUGGUGUACAAAUUAAUUAUU ........(((((....((((....(.((((((..----..---)))))).------).(((.....)))))))-...)))))............... ( -14.10) >consensus GAAAACCCGGCCAUUUAUGGCCAAAGCAUAAAACA_________UGCAGUGCCGCGGCCAAGCUCGAUUUGCCAAAAUUGGCCAACAAAUUAAUUAUU ........(((((....((((....((..................))...((....))............))))....)))))............... (-12.63 = -12.22 + -0.41)


| Location | 11,115,906 – 11,116,004 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 81.11 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11115906 98 - 27905053 AAUAAUUAAUUUGUUGGCCAAUUUUGGCAAAUCGAGCUUGGCCGCGGCACUGCGGCACUGCACUGUUUUAUGCUUUGGCCAUAAAUGGCCGGGUUUUC .............(((((((.((.((((.......((...((((((....))))))...))...((.....))....)))).)).)))))))...... ( -31.50) >DroPse_CAF1 31613 84 - 1 AAUAAUUAAUUUGUACACCAAAU-UGGCAAGUCGGGCUUUG------UACAGCA---UU----UGCGCUAUGCUUUGGCCAUAGAUGGUAAUGUUUUC .....((((((((.....)))))-)))...(((.((((..(------((.(((.---..----...))).)))...))))...)))............ ( -17.50) >DroSec_CAF1 26339 89 - 1 AAUAAUUAAUUUGUUGGCCAAUUUUGGCAAAUCGAGCUUGGCCGCGGCACUGCA---------UGUUUUAUGCUUUGACCAUAAAUGGCCGGGUUUUC ................((((....)))).....(((((((((((.((((..(((---------((...)))))..)).)).....))))))))))).. ( -25.70) >DroEre_CAF1 26792 89 - 1 AAUAAUUAAUUUGUUGGCCAAUUUUGGAAAAUCGAGCUUGGCCGCGGCACUGCA---------UGUUUUAUGCUUUGGCCAUAAAUGGCCGGGUUUUC ...........((((((((((.((((......)))).)))))))..)))..(((---------((...)))))((((((((....))))))))..... ( -24.80) >DroYak_CAF1 27255 89 - 1 AAUAAUUAAUUUGUUGGCCAAUUUUGGCAAAUCGAGCUUGGCCGCGGCACUGCA---------UGUUUUAUGCUUUGGCCAUAAAUGGCCGGGUUUUC ................((((....)))).....(((((((((((.(((...(((---------((...)))))....))).....))))))))))).. ( -27.40) >DroPer_CAF1 31730 84 - 1 AAUAAUUAAUUUGUACACCAAAU-UGGCAAGUCGGGCUUUG------UACAGCA---UU----UGCGCUAUGCUUUGGCCAUAGAUGGUAAUGUUUUC .....((((((((.....)))))-)))...(((.((((..(------((.(((.---..----...))).)))...))))...)))............ ( -17.50) >consensus AAUAAUUAAUUUGUUGGCCAAUUUUGGCAAAUCGAGCUUGGCCGCGGCACUGCA_________UGUUUUAUGCUUUGGCCAUAAAUGGCCGGGUUUUC ...............(((((.((.((((.....((((...((....))...((((.......)))).....))))..)))).)).)))))........ (-15.47 = -15.52 + 0.05)

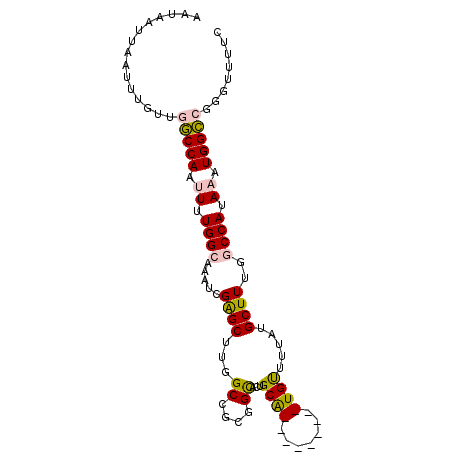

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:33 2006