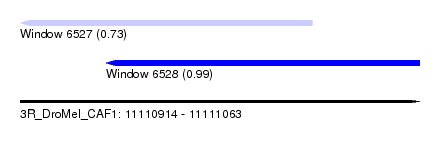
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,110,914 – 11,111,063 |
| Length | 149 |
| Max. P | 0.988538 |

| Location | 11,110,914 – 11,111,023 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -21.48 |
| Energy contribution | -22.77 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
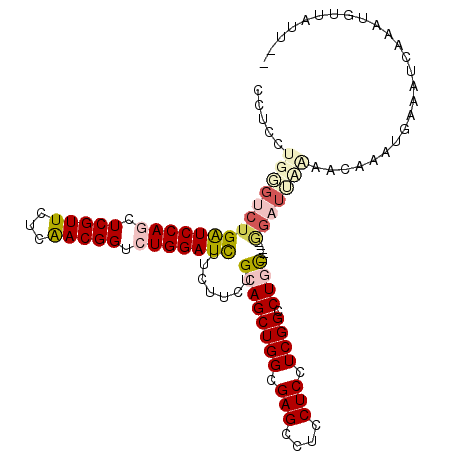
>3R_DroMel_CAF1 11110914 109 - 27905053 CUUCCUGGGUCUGAUCCAGCUCGUUCUCAACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUAC-----GGAUUCAAGCGAAUGAAAUCCAAUGUUAUUUG .....(((((..(((((((..(((.....)))..)))))))((((((.((.(((((.(((....))).)))))(...-----.)......))))).)))))))).......... ( -31.00) >DroGri_CAF1 24018 93 - 1 CCUCUUGUGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUUUUCUUUUGCAGCUGGCGAGCCUCCUCCUCGGCCUGC-----AGAUAAC-ACGAAAGAA--------------- .((.((((((..((((((..((((.....))))..))))))....(((((((((((.(((....))).)))).))))-----)))..))-)))).))..--------------- ( -30.60) >DroEre_CAF1 21726 108 - 1 CUUCCUGGGUCUGAUCCAGCUCGUUCUCAACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUGC-----GGAUUCGAGCAAAUGAAAUCAAAUGUUAUUC- .....((((((((((((((..(((.....)))..))))))).......((((((((.(((....))).)))).))))-----)))))))((((..((....))..))))....- ( -34.00) >DroWil_CAF1 24497 102 - 1 CCUCCUGUGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUUUUCUUUUGCAGCUGGCGAGCCUCCUCCUCGGCCUGU------ACA-ACAACAAUUGAAA----AUGAUAAAA- .......((((.((((((..((((.....))))..))))))......(((((((((.(((....))).)))).))))------)..-.............----..))))...- ( -22.20) >DroYak_CAF1 22161 108 - 1 CCUCCUGGGUCUGGUCCAGCUCGUUCUCGACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUGC-----GGAUAUGAACGAAUGAAAUCAAAUGUUAUUA- ((....))....(((((((..((((...))))..)))))))(((.(((((((((((.(((....))).)))).))))-----)))...)))......................- ( -33.20) >DroAna_CAF1 21439 110 - 1 CUUCCUGGGUCUGAUCCAGCUCGUUUUCAACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUAUUGGGUGGUUUUAAAGAAAAAUAAUAAAGAAUUA---- ((.((..(....(((((((..(((.....)))..)))))))..........(((((.(((....))).)))))...)..)).))..........................---- ( -26.70) >consensus CCUCCUGGGUCUGAUCCAGCUCGUUCUCAACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUGC_____GGAUUAAAACAAAUGAAAUCAAAUGUUAUU__ .....((((((((((((((.(((((...))))).))))))).......((((((((.(((....))).)))).)))).....)))))))......................... (-21.48 = -22.77 + 1.28)


| Location | 11,110,946 – 11,111,063 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.73 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -35.41 |
| Energy contribution | -34.30 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11110946 117 - 27905053 UUGUUCUUCUCCUCCAGCUUGCCGGUGACCAGGGUCAGGGCUUCCUGGGUCUGAUCCAGCUCGUUCUCAACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUAC--- ................((((((((((((((((((........)))).)))).(((((((..(((.....)))..)))))))..........))))))))))................--- ( -42.00) >DroVir_CAF1 24775 117 - 1 UUGUUCUUCUCCUCCAGCUUGCCAGUGACAAGGGUGAGCGCUUCCUGGGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUCUUCUUUUGCAGCUGGCGAGCCUCCUCCUCGGCCUAC--- ................(((((((((((((.((((........))))..))).((((((..((((.....))))..))))))..........))))))))))................--- ( -37.30) >DroGri_CAF1 24034 117 - 1 UUGUUCUUCUCCUCCAGCUUGCCAGUGACAAGGGUGAGAGCCUCUUGUGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUUUUCUUUUGCAGCUGGCGAGCCUCCUCCUCGGCCUGC--- ................((((((((((.((((((((....))).)))))....((((((..((((.....))))..))))))..........))))))))))................--- ( -38.70) >DroWil_CAF1 24522 117 - 1 UUGUUCUUCUCCUCCAGUUUGCCAGUGACAAGGGUGAGGGCCUCCUGUGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUUUUCUUUUGCAGCUGGCGAGCCUCCUCCUCGGCCUGU--- ..............(((.((((((((..((((((...(((((....).))))((((((..((((.....))))..))))))..))))))..))))))))(((........)))))).--- ( -34.80) >DroMoj_CAF1 25629 115 - 1 UUGUUCUUCUCUUCCAGUUUGCCAGUGACCAGGGUCAAAGCCUCUUGGGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUCUUCUUUUGCAGUUGGCGAGCCUCCUCCUCGGCCU----- ................(((((((((.((((.((((....))))....)))).((((((..((((.....))))..))))))...........)))))))))..............----- ( -32.40) >DroAna_CAF1 21469 120 - 1 UUGUUCUUCUCUUCAAGCUUGCCAGUGACCAGGGUCAGAGCUUCCUGGGUCUGAUCCAGCUCGUUUUCAACGGUCUGGAUCUUCUUCUGCAGCUGGCGAGCCUCCUCCUCGGCCUAUUGG ................((((((((((((((((((........)))).)))).(((((((..(((.....)))..)))))))..........))))))))))................... ( -42.70) >consensus UUGUUCUUCUCCUCCAGCUUGCCAGUGACAAGGGUCAGAGCCUCCUGGGUCUGAUCCAAUUCGUUUUCAACGGUCUGGAUCUUCUUUUGCAGCUGGCGAGCCUCCUCCUCGGCCUAC___ ................((((((((((((((((((........)))).)))).((((((..((((.....))))..))))))..........))))))))))................... (-35.41 = -34.30 + -1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:30 2006