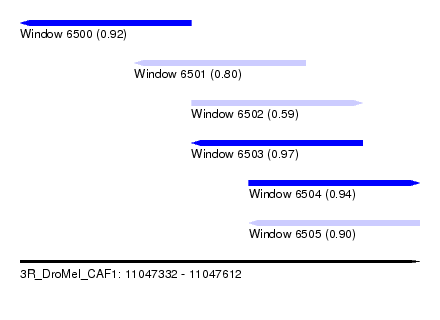
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,047,332 – 11,047,612 |
| Length | 280 |
| Max. P | 0.965467 |

| Location | 11,047,332 – 11,047,452 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -40.63 |
| Energy contribution | -40.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11047332 120 - 27905053 CAGCACCUCCAACCAGAGGUGAAGAACCUGUUCGUUGAGGCCCAAACAGAUCAGCGAGCGUAACUUAACAUCCAUUUGUGCGUGCGCAUCAUCGUGCGUCUGAUUGAUGGCCUGAACGCA ...((((((......))))))..(((....)))(((.((((((((.(((((..(((((((((.....(((......)))...)))))....))))..))))).)))..))))).)))... ( -42.10) >DroSec_CAF1 10102 120 - 1 CAGCACCUCCAACCACAGGUGAAGAACCUGUUCGUUGAGGCCCAAACAGAUCAGCGAGCGUAACUUAACAUCCAUUUGUGCGUGCGCAUCGUCGUGCGUCUGAUUGAUGGCCUGAACGCA ..((..........((((((.....))))))..(((.((((((((.((((((((((((((((.....(((......)))...))))).))))..)).))))).)))..))))).))))). ( -43.40) >DroSim_CAF1 10120 120 - 1 CAGCACCUCCAACCAGAGGUGAAGAACCUGUUCGUUGAGGCCCAAACAGAUCAGCGAGCGUAACUUAACAUCCAUUUGUGCGUGCGCAUCGUCGUGCGUCUGAUUGAUGGCCUGAACGCA ...((((((......))))))..(((....)))(((.((((((((.((((((((((((((((.....(((......)))...))))).))))..)).))))).)))..))))).)))... ( -43.20) >consensus CAGCACCUCCAACCAGAGGUGAAGAACCUGUUCGUUGAGGCCCAAACAGAUCAGCGAGCGUAACUUAACAUCCAUUUGUGCGUGCGCAUCGUCGUGCGUCUGAUUGAUGGCCUGAACGCA ...((((((......))))))..(((....)))(((.((((((((.(((((..(((((((((.....(((......)))...)))))....))))..))))).)))..))))).)))... (-40.63 = -40.97 + 0.33)


| Location | 11,047,412 – 11,047,532 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -39.57 |
| Energy contribution | -40.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11047412 120 - 27905053 CACUUGAUCUGCACCCAACCGGGUGAGUCAAUAAAGCUCCAGGAAUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCACAGCACCUCCAACCAGAGGUGAAGAACCUGUUCGUUGAGG ...(((((...(((((....))))).)))))........(((((.((....)).))))).....(((((..(((((..(....((((((......))))))..)..)))))..)..)))) ( -36.80) >DroSec_CAF1 10182 120 - 1 CACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCCAGGAGUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCACAGCACCUCCAACCACAGGUGAAGAACCUGUUCGUUGAGG ....((...(((.((..((((((.((((.......))))((((((((....))))))))........)))))).)).)))...))((((.(((.((((((.....))))))..))))))) ( -44.40) >DroSim_CAF1 10200 120 - 1 CACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCCAGGAGUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCACAGCACCUCCAACCAGAGGUGAAGAACCUGUUCGUUGAGG ...(((((...(((((....))))).)))))........((((((((....)))))))).....(((((..(((((..(....((((((......))))))..)..)))))..)..)))) ( -42.80) >consensus CACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCCAGGAGUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCACAGCACCUCCAACCAGAGGUGAAGAACCUGUUCGUUGAGG ...(((((...(((((....))))).)))))........((((((((....)))))))).....(((((..(((((..(....((((((......))))))..)..)))))..)..)))) (-39.57 = -40.23 + 0.67)

| Location | 11,047,452 – 11,047,572 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -43.53 |
| Consensus MFE | -41.21 |
| Energy contribution | -40.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11047452 120 + 27905053 UGCGCCUGCCAGGAGGUCGUACAGAAGUGGUAUCAUUCCUGGAGCUUUAUUGACUCACCCGGUUGGGUGCAGAUCAAGUGCGAGCUGCGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAAC ...((...(((((((...((((.......))))..))))))).))..........(((((....)))))(((((.(((.((((((((((........)))))))))))))..)))))... ( -45.80) >DroSec_CAF1 10222 120 + 1 UGCGCCUGCCAGGAGGUCGUACAGAAGUGGUAUCACUCCUGGAGCUUUAUUGACUCACCCGGCUGGGUGCAGAUCAAGUGCGAGCUACGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAAC .(((((((((.((........(((.((((....)))).)))((((......).))).)).))).))))))((((.(((.(((((((.((........)).))))))))))..)))).... ( -42.40) >DroSim_CAF1 10240 120 + 1 UGCGCCUGCCAGGAGGUCGUACAGAAGUGGUAUCACUCCUGGAGCUUUAUUGACUCACCCGGCUGGGUGCAGAUCAAGUGCGAGCUACGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAAC .(((((((((.((........(((.((((....)))).)))((((......).))).)).))).))))))((((.(((.(((((((.((........)).))))))))))..)))).... ( -42.40) >consensus UGCGCCUGCCAGGAGGUCGUACAGAAGUGGUAUCACUCCUGGAGCUUUAUUGACUCACCCGGCUGGGUGCAGAUCAAGUGCGAGCUACGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAAC .(((((((((.((........(((.((((....)))).)))((((......).))).)).))).))))))((((.(((.(((((((.((........)).))))))))))..)))).... (-41.21 = -40.77 + -0.44)


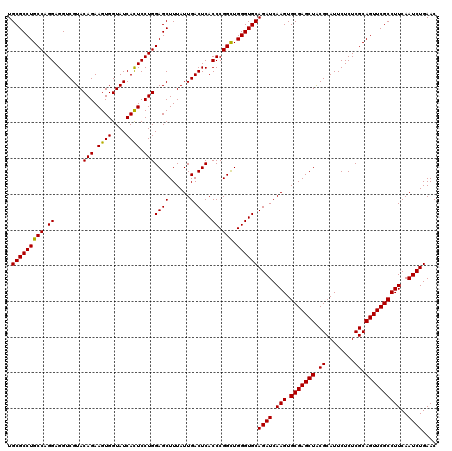
| Location | 11,047,452 – 11,047,572 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -45.83 |
| Consensus MFE | -44.85 |
| Energy contribution | -45.30 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11047452 120 - 27905053 GUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGCAGCUCGCACUUGAUCUGCACCCAACCGGGUGAGUCAAUAAAGCUCCAGGAAUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCA ...(((((..(..((((.(((((........))))).))))..)..)))))(((((....)))))..........(((((((((.((.(((.......)))))...))))))..)))... ( -42.50) >DroSec_CAF1 10222 120 - 1 GUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGUAGCUCGCACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCCAGGAGUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCA ....((((..(..((((.(((((........))))).))))..)..))))((.((..((((((.((((.......))))((((((((....))))))))........)))))).)).)). ( -47.50) >DroSim_CAF1 10240 120 - 1 GUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGUAGCUCGCACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCCAGGAGUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCA ....((((..(..((((.(((((........))))).))))..)..))))((.((..((((((.((((.......))))((((((((....))))))))........)))))).)).)). ( -47.50) >consensus GUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGUAGCUCGCACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCCAGGAGUGAUACCACUUCUGUACGACCUCCUGGCAGGCGCA ((.(((((..(..((((.(((((........))))).))))..)..))))).))((.((((((.((((.......))))((((((((....))))))))........)))))).)).... (-44.85 = -45.30 + 0.45)

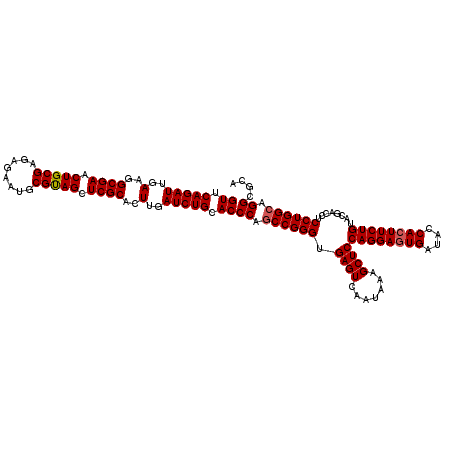
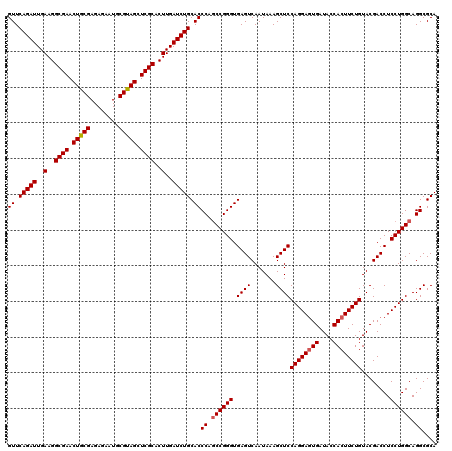
| Location | 11,047,492 – 11,047,612 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -52.37 |
| Consensus MFE | -46.24 |
| Energy contribution | -45.80 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11047492 120 + 27905053 GGAGCUUUAUUGACUCACCCGGUUGGGUGCAGAUCAAGUGCGAGCUGCGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAACCCCGACUGGGAACUGCCGCCGAAGCGCGGCAAGGAGUCGC ...........(((((.((((((((((..(((((.(((.((((((((((........)))))))))))))..)))))...))))))))))...((((((......))))))..))))).. ( -63.10) >DroSec_CAF1 10262 120 + 1 GGAGCUUUAUUGACUCACCCGGCUGGGUGCAGAUCAAGUGCGAGCUACGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAACCCCGACUGGGAACUGCCUCCGAAACGCGGCAAGGAGUCGC ...........(((((.((((((.((((.(((((.(((.(((((((.((........)).))))))))))..))))).)))).).)))))...((((.(......).))))..))))).. ( -47.30) >DroSim_CAF1 10280 120 + 1 GGAGCUUUAUUGACUCACCCGGCUGGGUGCAGAUCAAGUGCGAGCUACGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAACCCCGACUGGGAACUGCCCCCGAAACGCGGUAAGGAGUCGC ...........(((((.((((((.((((.(((((.(((.(((((((.((........)).))))))))))..))))).)))).).))))).(((((.........)))))...))))).. ( -46.70) >consensus GGAGCUUUAUUGACUCACCCGGCUGGGUGCAGAUCAAGUGCGAGCUACGCAUUCUCUCGCAGUUCGCCUUCAAUCUGAACCCCGACUGGGAACUGCCCCCGAAACGCGGCAAGGAGUCGC ...........(((((.((((((.((((.(((((.(((.(((((((.((........)).))))))))))..))))).)))).).)))))...((((.(......).))))..))))).. (-46.24 = -45.80 + -0.44)

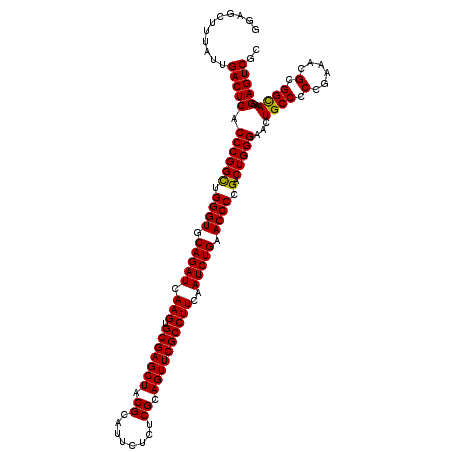
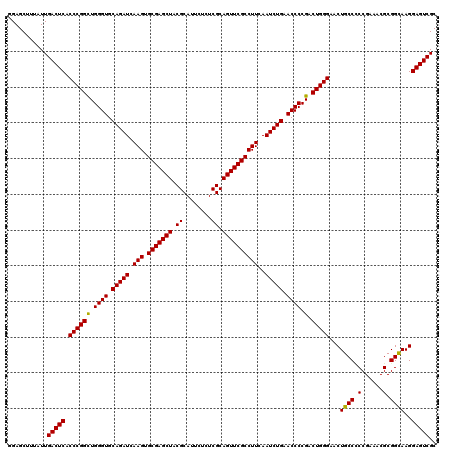
| Location | 11,047,492 – 11,047,612 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.94 |
| Mean single sequence MFE | -50.47 |
| Consensus MFE | -47.61 |
| Energy contribution | -47.50 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11047492 120 - 27905053 GCGACUCCUUGCCGCGCUUCGGCGGCAGUUCCCAGUCGGGGUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGCAGCUCGCACUUGAUCUGCACCCAACCGGGUGAGUCAAUAAAGCUCC ..(((((.(((((((......)))))))..(((.((..((((.(((((..(..((((.(((((........))))).))))..)..))))).)))).)).))).)))))........... ( -55.80) >DroSec_CAF1 10262 120 - 1 GCGACUCCUUGCCGCGUUUCGGAGGCAGUUCCCAGUCGGGGUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGUAGCUCGCACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCC ..(((((.(((((.(......).)))))..(((.((..((((.(((((..(..((((.(((((........))))).))))..)..))))).)))).)).))).)))))........... ( -48.40) >DroSim_CAF1 10280 120 - 1 GCGACUCCUUACCGCGUUUCGGGGGCAGUUCCCAGUCGGGGUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGUAGCUCGCACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCC ..(((((....(((.(((..((((.....)))).....((((.(((((..(..((((.(((((........))))).))))..)..))))).))))))))))..)))))........... ( -47.20) >consensus GCGACUCCUUGCCGCGUUUCGGAGGCAGUUCCCAGUCGGGGUUCAGAUUGAAGGCGAACUGCGAGAGAAUGCGUAGCUCGCACUUGAUCUGCACCCAGCCGGGUGAGUCAAUAAAGCUCC ..(((((.(((((.(......).)))))..(((.((..((((.(((((..(..((((.(((((........))))).))))..)..))))).)))).)).))).)))))........... (-47.61 = -47.50 + -0.11)

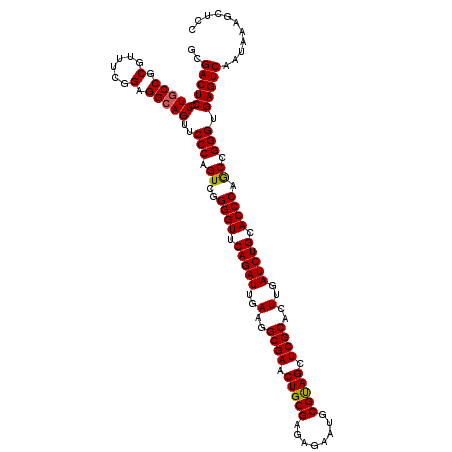
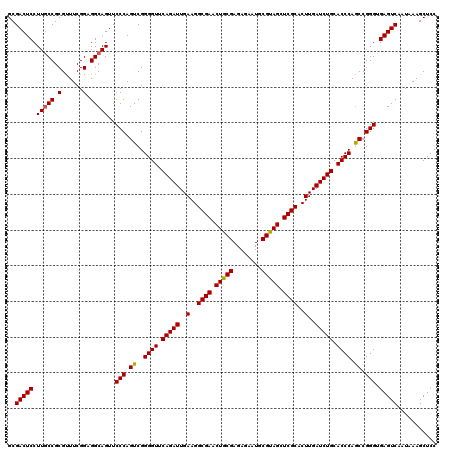
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:06 2006