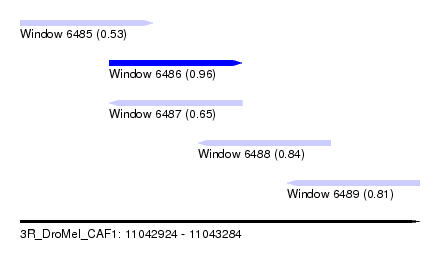
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 11,042,924 – 11,043,284 |
| Length | 360 |
| Max. P | 0.964502 |

| Location | 11,042,924 – 11,043,044 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -45.64 |
| Consensus MFE | -42.38 |
| Energy contribution | -43.28 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11042924 120 + 27905053 UUCCCAGGUCCAGGCCAACCAAACGGGCCGAUCGCAAGCGUGUCACAUGCUUGUACGUACUGAUGGCCAACUCUGCCAGCUCCAGGUAGAGCUUCUGGAAGGCUUGAUUCGAGCCCAGGG ..(((...(((((((((......(((..((...(((((((((...))))))))).))..))).)))))..(((((((.......)))))))....)))).((((((...))))))..))) ( -46.90) >DroSec_CAF1 5673 120 + 1 UUCCCAGGUCCAGACCAACCAAACGGGCGGAUCGCAAGCGUGUCACAUGCUUGUACGUACUGAUGGCCAGUUCUGCCAGCUCCAGGUAUAGCUUCUGAAAGGCUUGAUUCGAGCCCAGGG ..(((.(((....)))........((((((((((((((((((...)))))))))..(((((..((((.......))))......)))))(((((.....))))).)))))..)))).))) ( -40.20) >DroSim_CAF1 5693 120 + 1 UUCCCAGGUCCAGACCAACCAAACGGGCCGAUCGCAAGCGUGUCACAUGCUUGUACGUACUGAUGGCCAGUUCUGCCAGCUCCAGGUAGAGCUUCUGGAAGGCUUGGUUCGAGCCCAGGG ..(((...((((((...........((((.((((((((((((...))))))))).......)))))))(((((((((.......))))))))))))))).((((((...))))))..))) ( -49.81) >consensus UUCCCAGGUCCAGACCAACCAAACGGGCCGAUCGCAAGCGUGUCACAUGCUUGUACGUACUGAUGGCCAGUUCUGCCAGCUCCAGGUAGAGCUUCUGGAAGGCUUGAUUCGAGCCCAGGG ..(((...((((((...........((((.((((((((((((...))))))))).......)))))))(((((((((.......))))))))))))))).((((((...))))))..))) (-42.38 = -43.28 + 0.89)



| Location | 11,043,004 – 11,043,124 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -53.10 |
| Consensus MFE | -49.67 |
| Energy contribution | -50.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11043004 120 + 27905053 UCCAGGUAGAGCUUCUGGAAGGCUUGAUUCGAGCCCAGGGCUUCCUUAAGCUGCUCAGCCCCCGAUUUCUUGGGCUUACGAUUGGGAGACCGGUCUCGUAGCUGAGGUGUCGCCUGGAGG (((((((..(((((..(((((.((((.........)))).)))))..))))).((((((.(((((....))))).....((((((....)))))).....)))))).....))))))).. ( -55.80) >DroSec_CAF1 5753 120 + 1 UCCAGGUAUAGCUUCUGAAAGGCUUGAUUCGAGCCCAGGGCUUCCUUUAGCUGCUCAGCCCCCGAUUUUUUGGGCUUACGAUUGGGAGACCGUUCUCGGAGCUGAGGUGUCGCCUGGAGG (((((((..((((.(((...((((((...)))))))))))))..((((((((.(..(((((..........)))))...((.(((....))).))..).))))))))....))))))).. ( -48.20) >DroSim_CAF1 5773 120 + 1 UCCAGGUAGAGCUUCUGGAAGGCUUGGUUCGAGCCCAGGGCUUCCUUCAGCUGCUCAGCCCCCGAUUUUUUGGGCUUACGAUUGGGAGACCGUUCUCGGAGCUGAGGUGUCGCCUGGAGG (((((((..((((...(((((.(((((.......))))).)))))...)))).((((((.(((((....)))))....(((.(((....)))...)))..)))))).....))))))).. ( -55.30) >consensus UCCAGGUAGAGCUUCUGGAAGGCUUGAUUCGAGCCCAGGGCUUCCUUAAGCUGCUCAGCCCCCGAUUUUUUGGGCUUACGAUUGGGAGACCGUUCUCGGAGCUGAGGUGUCGCCUGGAGG (((((((..((((...(((((.((((.........)))).)))))...)))).((((((.(((((....)))))....(((.(((....)))...)))..)))))).....))))))).. (-49.67 = -50.00 + 0.33)



| Location | 11,043,004 – 11,043,124 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -40.93 |
| Consensus MFE | -38.44 |
| Energy contribution | -38.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11043004 120 - 27905053 CCUCCAGGCGACACCUCAGCUACGAGACCGGUCUCCCAAUCGUAAGCCCAAGAAAUCGGGGGCUGAGCAGCUUAAGGAAGCCCUGGGCUCGAAUCAAGCCUUCCAGAAGCUCUACCUGGA ..((((((......(((((((..((((....))))...........(((.(....).)))))))))).(((((..(((((.(.(((.......))).).)))))..)))))...)))))) ( -41.60) >DroSec_CAF1 5753 120 - 1 CCUCCAGGCGACACCUCAGCUCCGAGAACGGUCUCCCAAUCGUAAGCCCAAAAAAUCGGGGGCUGAGCAGCUAAAGGAAGCCCUGGGCUCGAAUCAAGCCUUUCAGAAGCUAUACCUGGA ..((((((......(((((((((((((....)))).....(....)............))))))))).((((..(((....)))(((((.......)))))......))))...)))))) ( -38.50) >DroSim_CAF1 5773 120 - 1 CCUCCAGGCGACACCUCAGCUCCGAGAACGGUCUCCCAAUCGUAAGCCCAAAAAAUCGGGGGCUGAGCAGCUGAAGGAAGCCCUGGGCUCGAACCAAGCCUUCCAGAAGCUCUACCUGGA ..((((((......(((((((((((((....)))).....(....)............))))))))).((((...(((((.(.(((.......))).).)))))...))))...)))))) ( -42.70) >consensus CCUCCAGGCGACACCUCAGCUCCGAGAACGGUCUCCCAAUCGUAAGCCCAAAAAAUCGGGGGCUGAGCAGCUAAAGGAAGCCCUGGGCUCGAAUCAAGCCUUCCAGAAGCUCUACCUGGA ..((((((......(((((((..((((....))))...........(((........)))))))))).((((...(((((.(.(((.......))).).)))))...))))...)))))) (-38.44 = -38.00 + -0.44)



| Location | 11,043,084 – 11,043,204 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -45.77 |
| Energy contribution | -46.10 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11043084 120 - 27905053 ACGCCAACUUCGGAGCAGUUGCACAUAGUGGUGCAGCUAUCGUCGGGAAUUGGAGAUGCGCCGUCCGAGCAGCAUCAGUUCCUCCAGGCGACACCUCAGCUACGAGACCGGUCUCCCAAU ..(((...((((.(((((((((((......)))))))).(((((.(((...(((((((((((....).)).)))))...)))))).))))).......))).))))...)))........ ( -41.20) >DroSec_CAF1 5833 120 - 1 ACGCCAACUUCGGAGCAGUUGCACAUAGUGGUGCAGCUAUCGUCGGGAAUUGGAGAUGCGCCGUCCGAGCAGCAUCAGUUCCUCCAGGCGACACCUCAGCUCCGAGAACGGUCUCCCAAU ..(((...((((((((((((((((......)))))))).(((((.(((...(((((((((((....).)).)))))...)))))).))))).......))))))))...)))........ ( -48.00) >DroSim_CAF1 5853 120 - 1 ACGCCAACUUCGGAGCAGUUGCACAUCGUGGUGCAGCUAUCGUCGGGAAUUGGAGAUGCGCCGUCCGAGCAGCAUCAGUUCCUCCAGGCGACACCUCAGCUCCGAGAACGGUCUCCCAAU ..(((...((((((((((((((((......)))))))).(((((.(((...(((((((((((....).)).)))))...)))))).))))).......))))))))...)))........ ( -48.00) >consensus ACGCCAACUUCGGAGCAGUUGCACAUAGUGGUGCAGCUAUCGUCGGGAAUUGGAGAUGCGCCGUCCGAGCAGCAUCAGUUCCUCCAGGCGACACCUCAGCUCCGAGAACGGUCUCCCAAU ..(((...((((((((((((((((......)))))))).(((((.(((...(((((((((((....).)).)))))...)))))).))))).......))))))))...)))........ (-45.77 = -46.10 + 0.33)



| Location | 11,043,164 – 11,043,284 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -45.50 |
| Consensus MFE | -45.50 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 11043164 120 - 27905053 AGCACUGUGCUCCCAUCUGGAACUUGGCCAAGGAUAAGCUCUACGAGCUCGGCAAACUGUGUGGCCUCUUGCCCGGCUGCACGCCAACUUCGGAGCAGUUGCACAUAGUGGUGCAGCUAU ........((((((....)(((.(((((....(...(((((...)))))...).....(((..(((........)))..)))))))).))))))))((((((((......)))))))).. ( -45.50) >DroSec_CAF1 5913 120 - 1 AGCACUGUGCUCCCAUCUGGAACUUGGCCAAGGAUAAGCUCUACGAGCUCGGCAAACUGUGUGGCCUCUUGCCCGGCUGCACGCCAACUUCGGAGCAGUUGCACAUAGUGGUGCAGCUAU ........((((((....)(((.(((((....(...(((((...)))))...).....(((..(((........)))..)))))))).))))))))((((((((......)))))))).. ( -45.50) >DroSim_CAF1 5933 120 - 1 AGCACUGUGCUCCCAUCUGGAACUUGGCCAAGGAUAAGCUCUACGAGCUCGGCAAACUGUGUGGCCUCUUGCCCGGCUGCACGCCAACUUCGGAGCAGUUGCACAUCGUGGUGCAGCUAU ........((((((....)(((.(((((....(...(((((...)))))...).....(((..(((........)))..)))))))).))))))))((((((((......)))))))).. ( -45.50) >consensus AGCACUGUGCUCCCAUCUGGAACUUGGCCAAGGAUAAGCUCUACGAGCUCGGCAAACUGUGUGGCCUCUUGCCCGGCUGCACGCCAACUUCGGAGCAGUUGCACAUAGUGGUGCAGCUAU ........((((((....)(((.(((((....(...(((((...)))))...).....(((..(((........)))..)))))))).))))))))((((((((......)))))))).. (-45.50 = -45.50 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:49 2006